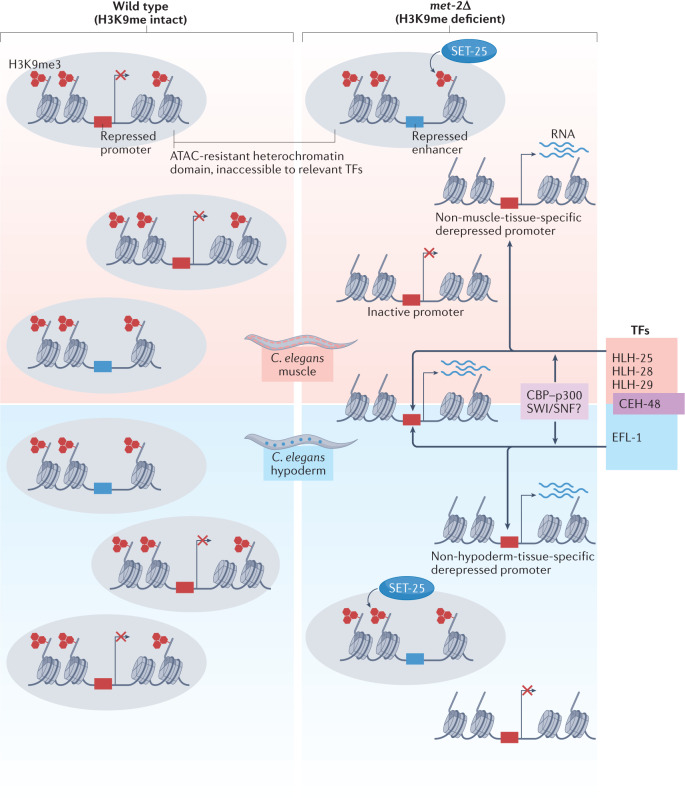
Fig. 4. Transcription factors control tissue-specific transcription derepression upon H3K9me loss.
A major question is whether histone H3 Lys9 (H3K9) methylation contributes to gene repression in differentiated tissues by blocking transcription factor (TF) binding to chromatin218. A recent study addressed this question in Caenorhabditis elegans83 by comparing the effects of loss of the SET domain bifurcated 1 (SETDB1)-like histone methyltransferase MET-2 on gene expression in embryos and in isolated muscle and hypoderm tissues. As depicted schematically, the genes upregulated by MET-2 loss constitute only a fraction (12% in embryos and 13% in muscle) of the genes marked by trimethylated H3K9 (H3K9me3), and the subset of genes affected is both tissue specific and developmental stage specific. In muscle and hypoderm, H3K9 methylation by MET-2 represses tissue-specific genes expressed in other cell lineages, including, but not restricted to, germline-specific genes83,219,220. In addition, MET-2 and SET-25 redundantly repress a large number of enhancer elements in muscle. Methylated H3K9 (H3K9me) heterochromatin ensures transcription inactivity by restricting the binding of a defined set of transcription factors at promoters and enhancers, which are either tissue specific — for example, HLH25, HLH-28 and HLH-29 in muscle and EFL-1 in hypoderm — or more widely expressed, for example CEH-48 (ref.83). As measured by assay for transposase-accessible chromatin with high-throughput sequencing (ATAC–seq), increased accessibility upon loss of H3K9me is neither sufficient nor necessary to drive transcription, but a subset of β-helix–turn–helix and bZip-containing factors that can recruit chromatin remodelling complexes such as SWI/SNF or the acetyltransferase complex CBP–p300 drive transcription at the genes that become both accessible and upregulated83. These results suggest that H3K9 methylation confers tissue-specific gene expression by restricting transcription factor access. The met-2 mutation compromises both muscle ultrastructure and worm mobility.