Extended Data Fig.8 ∣. BRD2 interplays with Cohesin. Related to Fig.3.
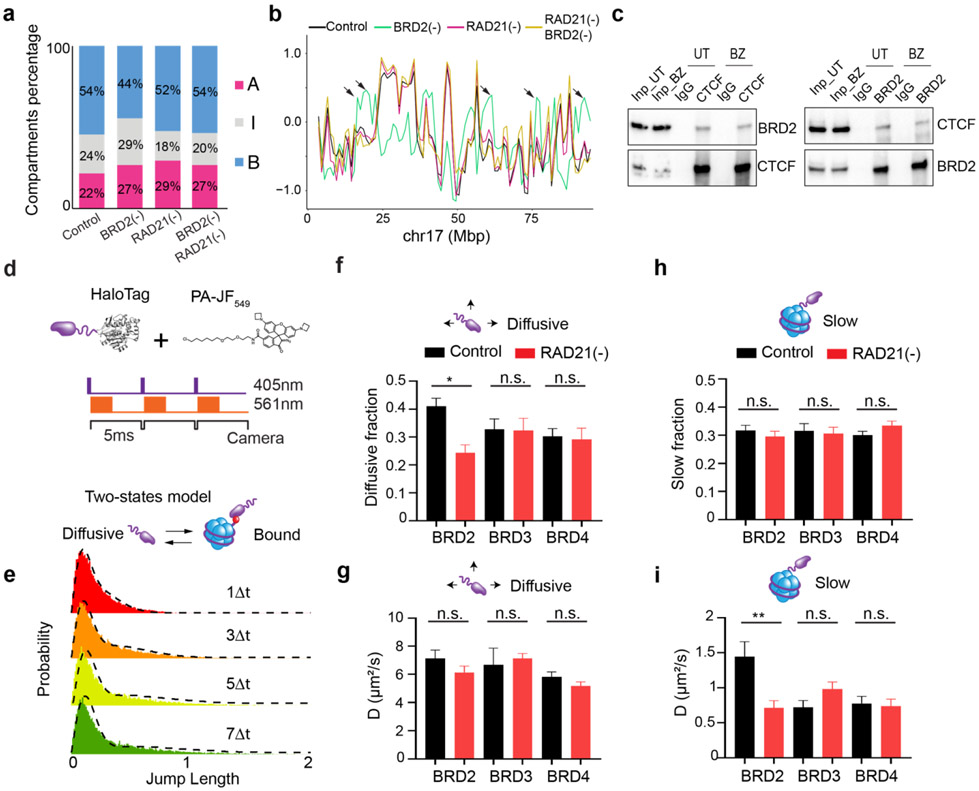
(a) Quantification of compartmental changes after BRD2 and RAD21 depletion individually or in combination. A, active compartments; I, intermediated mixed compartments; B, inactive compartments. The mouse genome is binned into 25480 segments (1Mbp bin size) and the percentage of each fraction is shown. See details of compartment analysis in the method section.
(b) Browser track view of the eigenvector values for compartmental score in various perturbation conditions on chromosome 17. The genomic regions containing B to A switches after BRD2 depletion (green line) are highlighted with black arrows.
(c) Endogenous Co-immunoprecipitation between BRD2 and CTCF in mouse ESCs. Benzonase (BZ) treatment did not abrogate the BRD2-CTCF interaction as compared to untreated control (UT).
(d) (Upper panel) Schematic of genome engineering of BET proteins with HaloTag and labeled with Janelia photoactivatable fluorophore PA-JF549 for single molecule tracking. (Lower panel) Illustration of stroboscopic single molecule imaging of BET protein dynamics in live cells. After activation by a 405 nm pulse, single BET molecules were excited by 561 nm laser pulse (2 ms) to suppress motion blurring and images were captured with ~5ms exposure times.
(e) The probability distribution function of jump length was fit over multiple camera integration time scales by the two-state model estimated by Spot-On program (See methods). The two-state diffusive vs bound model does not well account for the dynamics of BRD2 or BRD3/4 (not shown) as compared to the three-state model in Fig.3c.
(f-g) The diffusive fraction (f) and its diffusion coefficient (g) analysis for BRD2, BRD3 and BRD4 from SMT analysis after Cohesin depletion. Cohesin depletion significantly decreased the diffusive fraction of BRD2 accompanied with an increase of the chromatin bound fraction. The number of cells analyzed are the same as Fig.3d-e.
(h-i) The slow bound fraction (h) and its diffusion coefficient (i) analysis for BRD2, BRD3 and BRD4 from SMT analysis after Cohesin depletion. Cohesin depletion also significantly decreased the diffusion coefficient of slow bound BRD2. The number of cells analyzed are the same as Fig.3d-e.