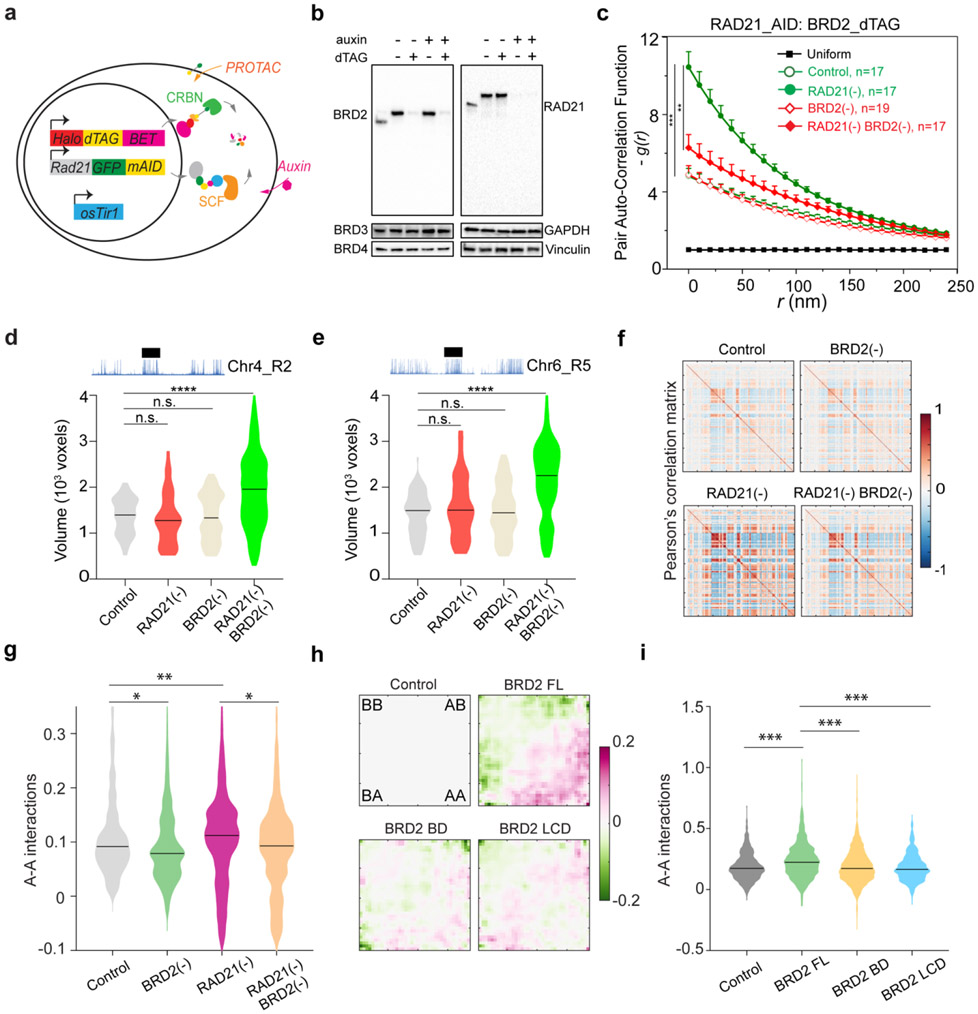
Fig.2 ∣. BRD2 mixes and compacts active compartments in the absence of Cohesin.
(a) A schematic of the dual targeted protein degradation strategy. In Rad21-eGFP-mAID cells, a FKBPF36V based degron (dTAG) linked to a HaloTag was bi-allelically knocked into endogenous BET family genes (Brd2, Brd3 and Brd4) by the CRISPR/Cas9 genome editing method (only one engineered allele is shown). Adding cell membrane permeable dTAG13 ligand into the culture will bring the dTAG labeled BET proteins into close proximity to the Cereblon (CRBN) E3 ligase for proteasome-mediated protein degradation orthogonal to the mAID system.
(b) Western blot (WB) analysis of protein levels of endogenous BRD2-dTAG or RAD21-AID individually or in combination after 6 hours of dTAG13 or auxin treatment, respectively. Rapid depletion of BRD2 or RAD21 individually or together does not impact the protein level of other BET proteins (BRD3 or BRD4).
(c) Degradation of BRD2 for 6 hours significantly reduced the accessible chromatin clustering after RAD21 depletion. g(r) curves were plotted for indicated conditions.
(d-e) Violin plot of 3D volumes (the number of voxels) of two ATAC-rich segments R2 (d) and R5 (e) before and after BRD2 depletion, RAD21 depletion or in combination. The black bar indicates the median value for each data set and Mann-Whitney U test was performed for the statistical analysis. The number of analyzed alleles for the ATAC-rich segment (Chr4-R2) are : Control (n=93), RAD21 depletion (n=129), BRD2 depletion (n=116), or dual depletion of both RAD21 and BRD2 (n=119). The number of analyzed alleles for the ATAC-rich segment (Chr4-R5) are : Control (n=91), RAD21 depletion (n= 85), BRD2 depletion (n=99), or dual depletion of both RAD21 and BRD2 (n=121).
(f) Pearson’s correlation matrix of the whole chromosome 17 for BRD2 or RAD21 depletion alone or in combination for 6 hours from Micro-C experiments. Dual BRD2 and RAD21 depletion reduced the enhanced compartmentalization after Cohesin depletion alone.
(g) Quantification of the digitalized A-A compartmental interactions (log10 value of observed/expected) after BRD2 or RAD21 depletion alone or in combination from Micro-C experiments. The black solid line in each violin plot represents the median value.
(h-i) Differential saddle plot analysis (h) and quantitative A-A compartmental interactions (log10 value of observed/expected) analysis (i) by Micro-C for cells stably expressing empty vector, full length (FL), N-terminal double bromodomain (BD) and C-terminal low complexity domain (LCD) of BRD2.
The non-parametric two-sided Mann-Whitney U test was used for statistical testing. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.