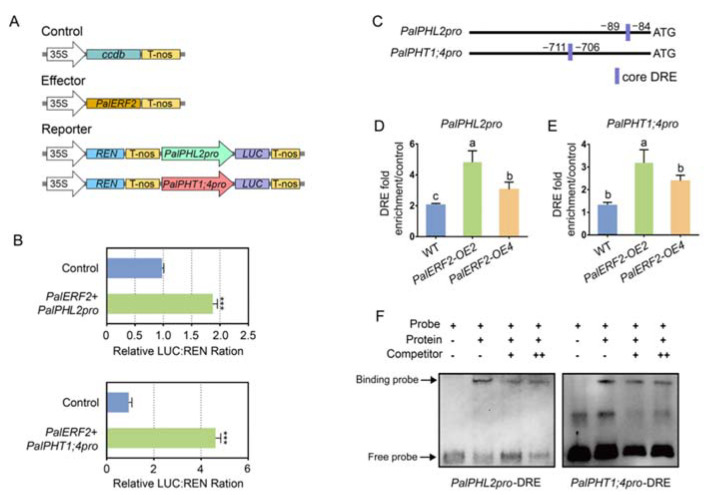
Figure 3.
PalERF2 regulates PalPHL2 and PalPHT1;4 expression. (A) Structures of effector and reporters employed in dual-luciferase assay. (B) Transient co-expression of effector and reporter vectors in Nicotiana benthamiana leaves for dual-luciferase assay. Error bars indicate SD values (n = 3). Asterisks indicate significant differences compared to control by Student’s t-test, ***, p < 0.01. (C) Distribution of core DRE motifs in the promoters of PalPHL2 and PalPHT1;4. (D,E) ChIP-qPCR determined the binding of PalERF2 to the PalPHL2 and PalPHT1;4 promoter regions containing DRE, respectively. Error values represent means ± SD (n = 3). Significant differences were analyzed by Duncan’s test (p < 0.05, n = 5). Different letters indicate statistically significant differences. (F) EMSA tested the binding activity of PalERF2 to the DRE in PalPHL2 and PalPHT1;4 promoters. The unlabeled cold probes were added to compete with labeled probes. + means the cold probe is 20 times the labeled probe, ++ means 50 times. The arrows mark the binding probe and free probe.