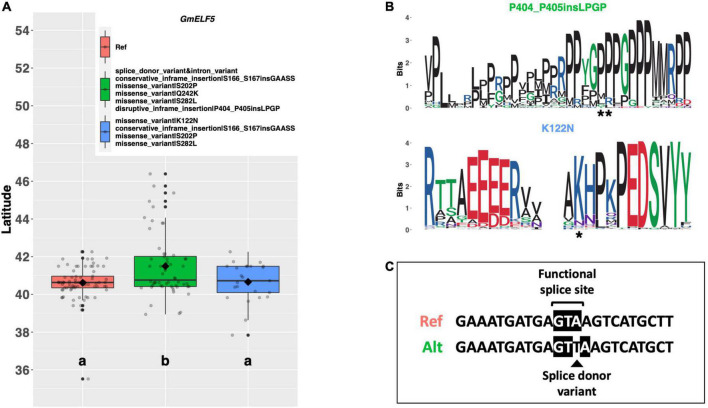
FIGURE 5.
(A) Latitudinal distribution of GmELF5 alleles among 187 resequenced ELs with the maturity genotype e1-as/E2. Latitude of origination was used a proxy for relative flowering time. Latitude values were estimated based on state of origin and, where available, were scaled according to maturity info provided by breeders that developed the ELs (see Experimental Procedures). Means comparison was conducted using an ANOVA, where the f-statistic representing the 95% confidence interval was empirically derived by randomization. Significance letters were obtained from a test of least significant difference. Transparent dots represent the latitude of each accession. Boxplots show the mean (diamond), median (solid line), quartile span (box), range (vertical lines), and outliers (solid dots). Inset legend shows the collection of mutations which make up each allele, where “Ref” refers to the Wm82.a2.v1 reference allele. (B) Weblogo depicting degree of amino acid conservation of the domain surrounding prioritized mutations (asterisk) in GmELF5. (C) Comparison of the splice site sequence at the second exon/intron boundary in the Williams 82 Ref sequence and alternate allele containing the splice donor variant.