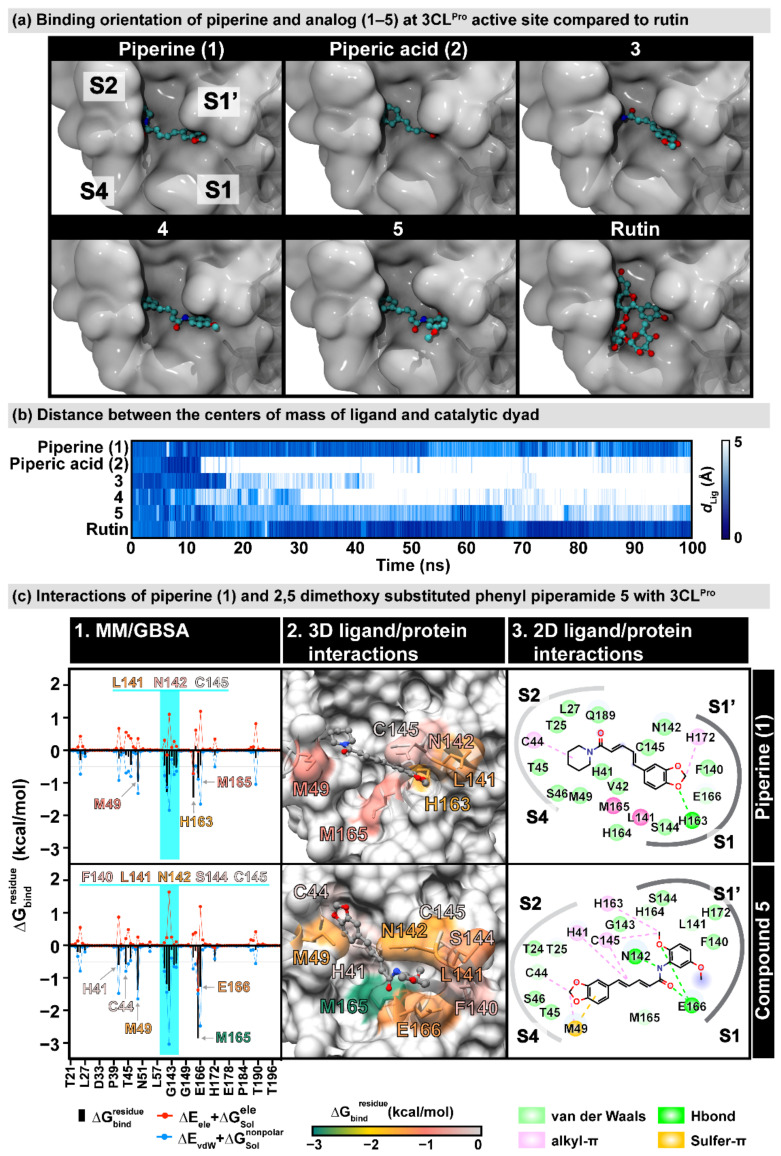
Figure 4.
(a) Predicted binding orientation of piperine and analogs (1–5) at the active site of SARS-CoV-2 3C-like (3CLPro) main protease obtained from molecular docking study compared to rutin [36]. (b) Distance between the center of the mass of the ligand and the catalytic dyad (H41 and C145) along with 100-ns molecular dynamics simulation of piperine and analogs (1–5). The colors are shaded from dark blue to white, indicating distances of 0 Å and 5 Å, respectively. (c) 3CLpro interactions of piperine (1) and 2,5-dimethoxy substituted phenyl piperamide 5. (c-1). MM/GBSA per-residue decomposition free energy () and its polar and nonpolar components of piperine (1) and 2,5-dimethoxy substituted phenyl piperamide (5). Residues with ≤ −0.5 kcal/mol are labeled, and the residues 140–145 in the oxyanion region are highlighted. (c-2). The 3D ligand-protein interactions are also shown according to values. (c-3). 2D ligand–protein interactions were drawn from the representative structure using BIOVIA Discovery Studio Visualizer V21.1.0.