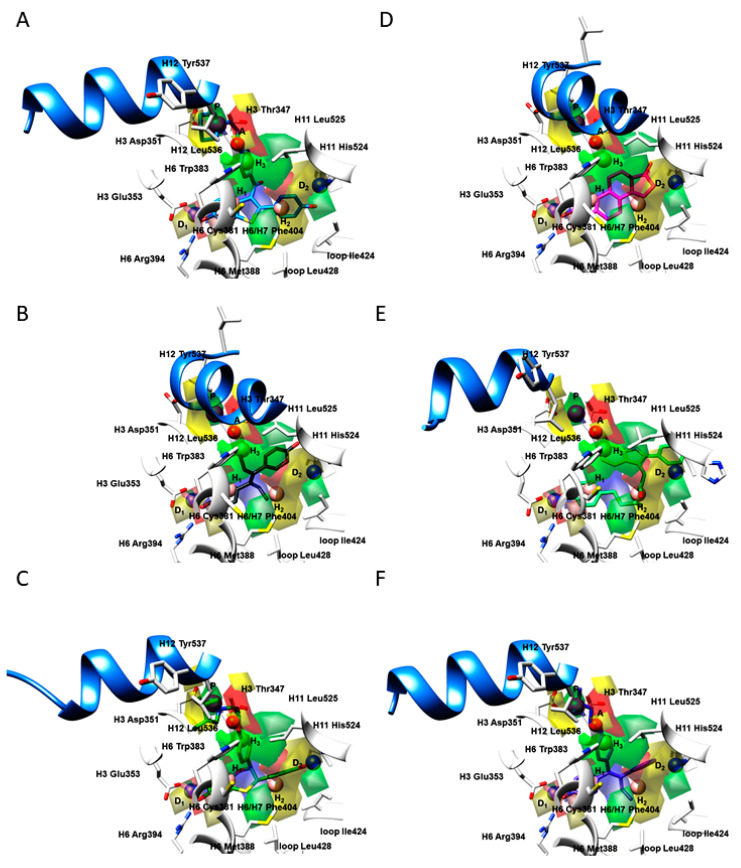
Figure 5.
The 3-D PhypI features (D: hydrogen-bond donators, A: hydrogen-bond acceptors, H: hydrophobic features, P: positive ionizable features) and 3-D QSAR PLS-coefficients contour maps (GREENPLS-coefficients: positive steric interactions, YELLOWPLS-coefficients; negative steric interactions, BLUEPLS-coefficients: areas where positively charged functional groups and H-bond donators are favored whereas the negatively charged functional groups and H-bond acceptors are disfavored, REDPLS-coefficients: areas negatively charged functional groups and H-bond acceptors are favored, whereas the positively charged functional groups and H-bond donators are disfavored) for 1ERR (A); 3ERD (B); 1XP1 (C); 1ERE (D); 2IOK (E); 2BJ4 (F). Amino acid residues are depicted in white. For the clarity of presentation, only the H12 helix is presented in a cornflower blue ribbon, as a crucial delimiter for partial agonists, SERMs, and SERDs.