Table 6.
Structures of designed hits and their predicted activities against ERα.
| # | Ligand Structure |
3DPhypI/3-D QSAR pred. pIC50 | # | Ligand Structure |
3DPhypI/3-D QSAR pred. pIC50 b | ||
|---|---|---|---|---|---|---|---|
| SB a | LB b | SB a | LB b | ||||
| 3DPQ-1 |
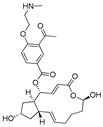
|
9.20 | 9.17 | 3DPQ-7 |
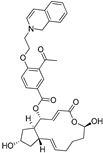
|
9.26 | 9.11 |
| 3DPQ-2 |
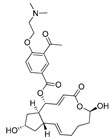
|
9.21 | 9.12 | 3DPQ-8 |
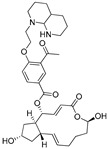
|
9.04 | 8.95 |
| 3DPQ-3 |
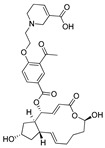
|
9.37 | 9.29 | 3DPQ-9 |
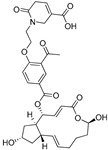
|
9.31 | 9.26 |
| 3DPQ-4 |
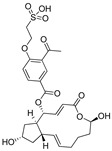
|
9.26 | 9.22 | 3DPQ-10 |
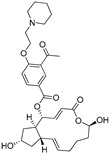
|
9.18 | 9.05 |
| 3DPQ-5 |
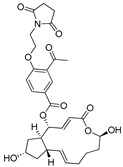
|
9.05 | 8.92 | 3DPQ-11 |
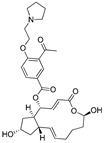
|
9.12 | 9.28 |
| 3DPQ-6 |
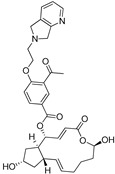
|
9.01 | 8.91 | 3DPQ-12 |
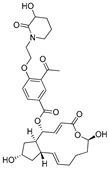
|
9.42 | 9.35 |
a The designed compounds SB predicted activities by the 3DPhypI/3-D QSAR model; b The designed compounds LB predicted activities by the 3DPhypI/3-D QSAR model.
