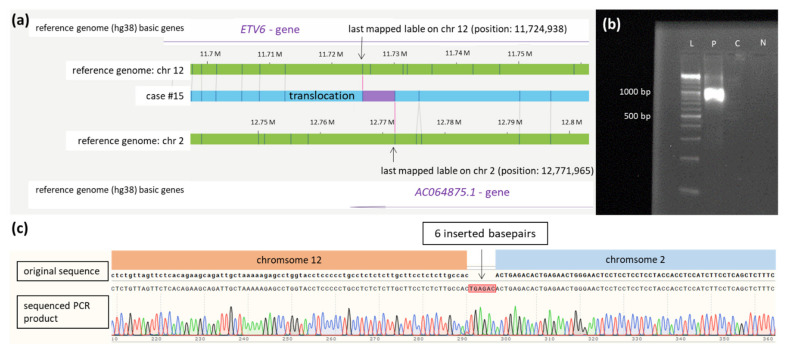
Figure 3.
Translocation t(2;12). (a) Schematic representation of the newly identified translocation t(2,12) in case #15 detected by OGM. The first bar represents the reference genome (hg38) basic genes of chromosome 12 located at the depicted position (ETV6). The second and fourth bars (green) show the reference genome map of chromosome 2 and chromosome 12. The third bar (blue) represents the map of case #15. The fifth bar represents the reference genome (hg38) basic genes of chromosome 2 located at the depicted position (AC064875.1). Vertical blue lines represent mapped labels. (b) Validation of the translocation t(2;12) in case #15 via PCR showing a product of 1138 bp spanning translocation t(2,12) (ranging from 12,767,669 on chromosome 2 to 11,725,810 on chromosome 12). The healthy control-DNA and the negative control were both negative in the PCR. (c) Sequenced PCR product spanning the breakpoint of translocation t(2;12) in case #15 with insertion of 6 base pairs. In the first row, the original sequence of chromosome 12 followed by chromosome 2 is shown (modified from the figure as shown in the SnapGene programTM). The second row depicts the electropherogram and sequence of PCR product of case #15 aligned to the original sequences spanning the breakpoint of the translocation. Insertion of 6 base pairs (marked red) at the breakpoint is shown in the middle of the second row and probably results from DNA repair mechanisms at the breakpoint. Abbreviations: C = control-DNA; chr = chromosome; hg38 = human genome project 38; ETV = ETS variant transcription factor 6; L = length marker; N = negative control (H2O); P = sample from case #9 collected at diagnosis.