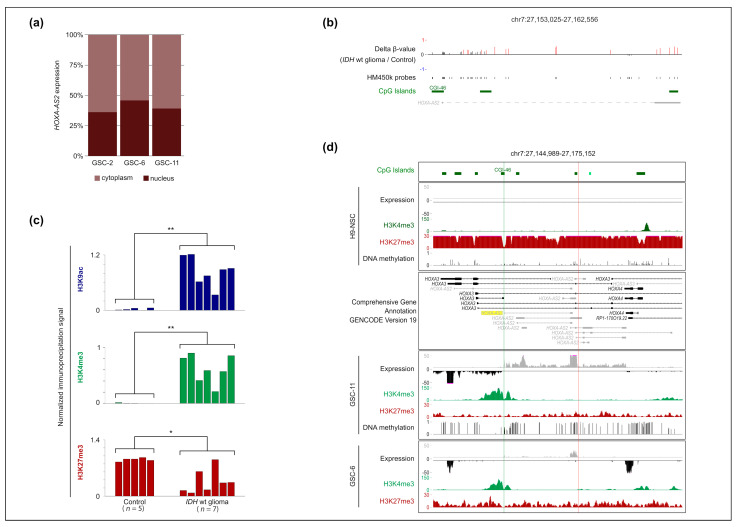
Figure 2.
Subcellular distribution and molecular profile of HOXA-AS2. (a) Relative distribution of HOXA-AS2 transcripts in cytoplasm and nucleus of three GSC lines. Expression levels were normalized to U6 and 18S expression in the cytoplasmic and nuclear fractions, respectively. (b) DNA methylation changes (compared with control samples; n = 5) along the HOXA-AS2 genomic region in IDHwt (n = 55) glioma samples, detected with the HM450K array. Significantly hypermethylated regions are in red (no hypomethylated region was observed). * p < 0.05, ** p < 0.01 (Mann-Whitney U-test). (c) ChIP analysis of H3K9ac, H3K4me3, and H3K27me3 at the HOXA-AS2 promoter region in control brain samples (n = 5), and IDHmut (n = 5) and IDHwt (n = 7) glioma samples. The values obtained for each sample are shown. The precipitation level was normalized to that at the TBP promoter (for H3K4me3 and H3K9ac) and at the SP6 promoter (for H3K27me3). (d) Genome Browser integrative view at the HOXA-AS2 locus to show H3K4me3, H3K27ac, and H3K27me3 enrichment, DNA methylation (in the GSC-11 and H9-NSC lines), and the strand-oriented RNA-seq signal in GSC-6, GSC-11, and H9-NSC lines. The positions of the 5′ and 3′ ends identified by RACE-PCR are shown by a green and red line, respectively.