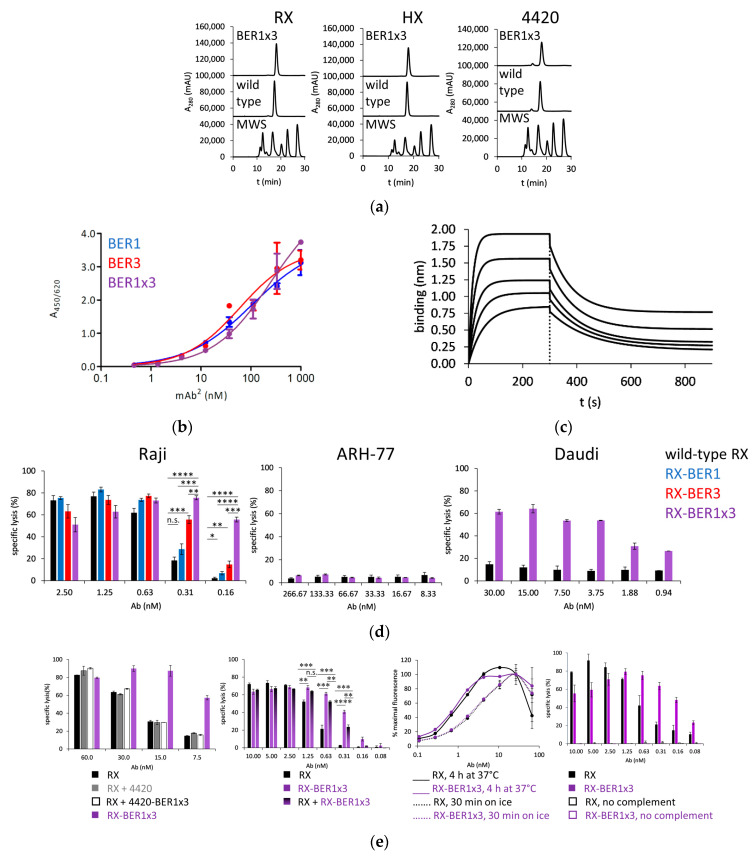
Figure 4.
Characterization of shuffled BER1x3-clone based mAb2 molecules: (a) SEC-HPLC profiles in native conditions of rituximab (RX)-, HuMax-20 (HX)- and 4420-based mAb2 molecules (MWS: molecular weight standard); (b) ELISA response of RX-based bispecific antibodies with parental Fcabs and the shuffled clone to immobilized CD59; (c) Biolayer interferometry-determined binding of RX-BER1x3 to immobilized CD59, antibody in 2-fold dilutions starting with 300 nM; (d) Complement-dependent cytotoxicity (CDC) effect of RX-BER1, -3 and -1x3 for Raji and RX-BER1x3 for ARH-77 and Daudi cells. Sample size was n = 3 for Raji and n = 2 for ARH-77 and Daudi cells. One-way ANOVA was used to determine the significance (n.s. (not significant) 0.05 ≤ p, * 0.01 ≤ p < 0.05, ** 0.001 ≤ p < 0.01, *** 0.0001 ≤ p < 0.001, **** p ≤ 0.0001), mean and S.D. are presented; (e) 1st panel: CDC effect on Raji cells of RX, mix of RX and 4420 antibody, mix of RX and 4420-BER1x3 mAb2 and RX-BER1x3 mAb2 (n = 2), 2nd panel: CDC effect on Raji cells of RX, RX-BER1x3, and mix of RX and RX-BER1x3 (n = 3). One-way ANOVA was used to determine the significance (n.s. (not significant) 0.05 ≤ p, ** 0.001 ≤ p < 0.01, *** 0.0001 ≤ p < 0.001, **** p ≤ 0.0001), 3rd panel: Raji cell staining with RX and RX-BER1x3 at 4 °C and at 37 °C (n = 2), 4th panel: the effect of RX and RX-BER1x3 on Raji cells with and without complement (n = 2). Mean and S.D. are presented.