Figure 4.
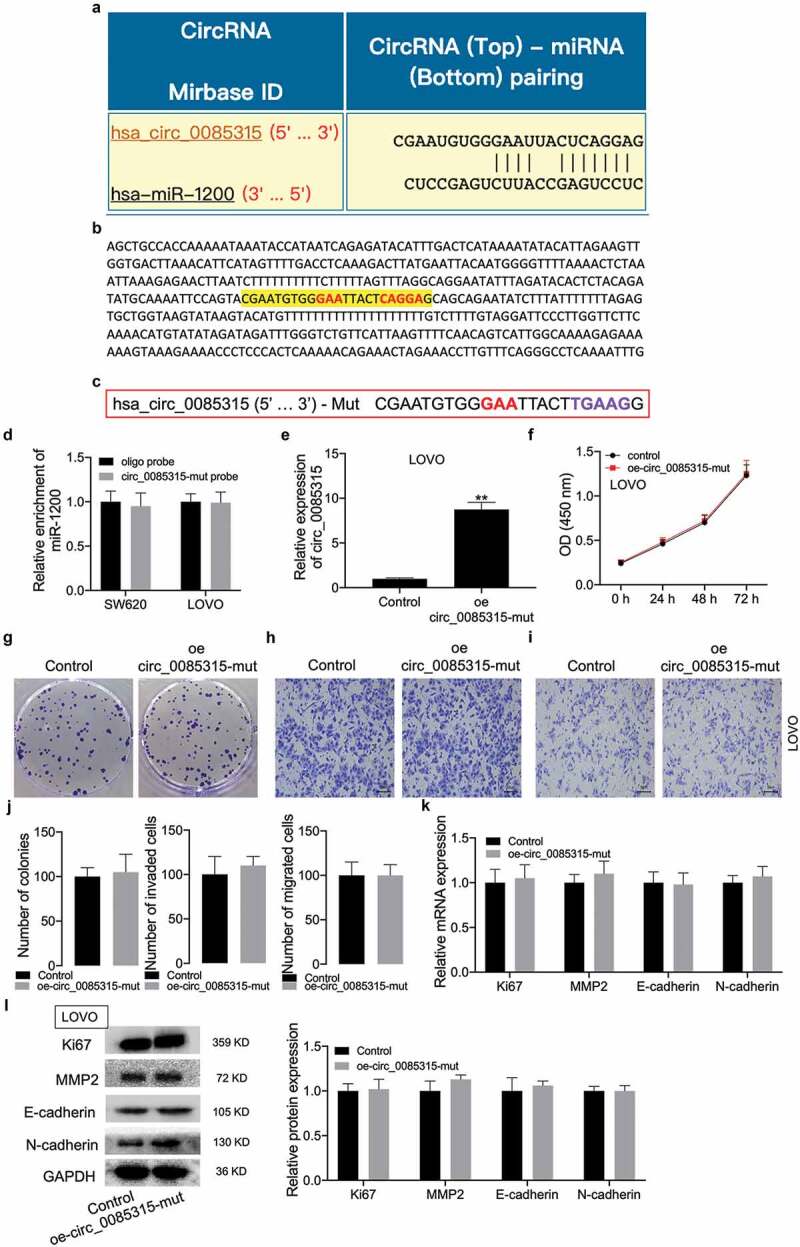
CircRNA_0085315 mut had no affect cell proliferation, invasion, and migration in CC.
(A) The binding sequence of miR-1200 on circRNA_0085315 is predicted from the Circular RNA Interactome database. (B) the site that can bind to miR-1200 locating on the full-length sequence of circ_0085315. (C) The sequence of circRNA_0085315 mut. (D) qRT-PCR assay was used to determine that the biotinylated circRNA_0085315-mut probe had no effect on the enrichment level of miR-1200. A vector expressing circRNA_0085315 mut was transfected into LOVO cells, circRNA_0085315 level (E), cell viability (F), proliferation (G), invasion (H), and migration (I) were detected by qRT-PCR, CCK-8, colony formation, Transwell assays. The statistical diagrams were shown in J. qRT-PCR assay (K) and Western blot assay (L) aimed at analyzing the expression of Ki67, MMP2, E-cadherin, and N-cadherin in CC cells under the transfection of oe-circ_0085315 mut. *P < 0.05; **P < 0.01.
