Figure 5.
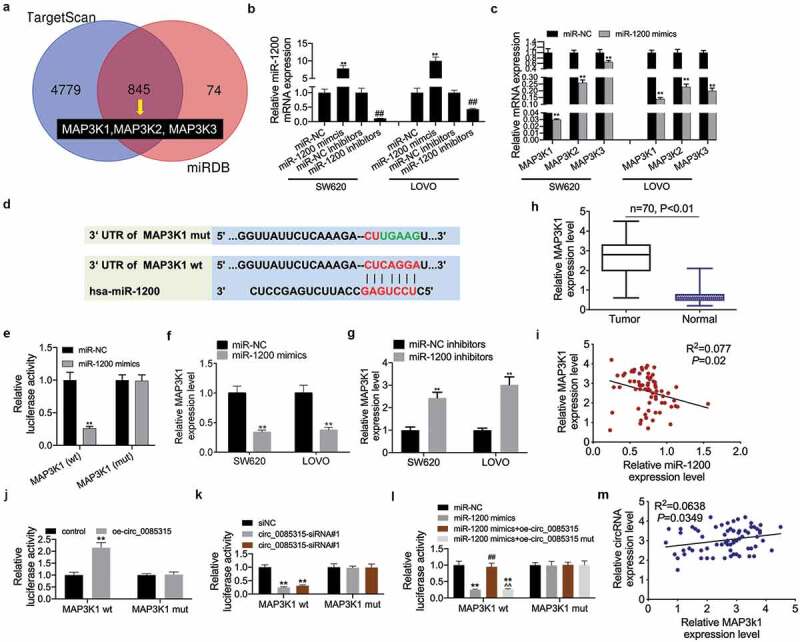
CircRNA_0085315 sponges to miR-1200 to upregulate MAP3K1 expression in CC cells.
(A) Venn analysis of 845 overlapping target genes between TargetScan and miRDB database. (B) The transfection efficiency of miR-1200 mimics or inhibitors was verified using qRT-PCR. (C) The expression of MAP3K1, MAP3K2, and MAP3K3 was detected using qRT-PCR in SW620 and LOVO cells transfected by miR-1200 mimics. (D) The binding sequence of miR-1200 on MAP3K1 is predicted from TargetScan. (E) The interaction between MAP3K1 and miR-1200 was assessed by luciferase reporter assay. (F and G) The expression of MAP3K1 was detected using qRT-PCR in SW620 and LOVO cells transfected by miR-1200 mimics or miR-1200 inhibitors. (H) Relative MAP3K1 expression in CC tumor tissues compared with adjacent normal tissues (n = 43) by qRT-PCR analysis. (I) Pearson correlation analysis of between MAP3K1 and miR-1200. (J and K) The interaction between MAP3K1 and circ_0085315 was assessed by luciferase reporter assay. (L) The interaction among MAP3K1, miR-1200, and circ_0085315 was assessed by luciferase reporter assay. *P < 0.05; **P < 0.01.
