Fig. 1 |. Overview of nucleobase reaction chemistry underlying ac4C-seq.
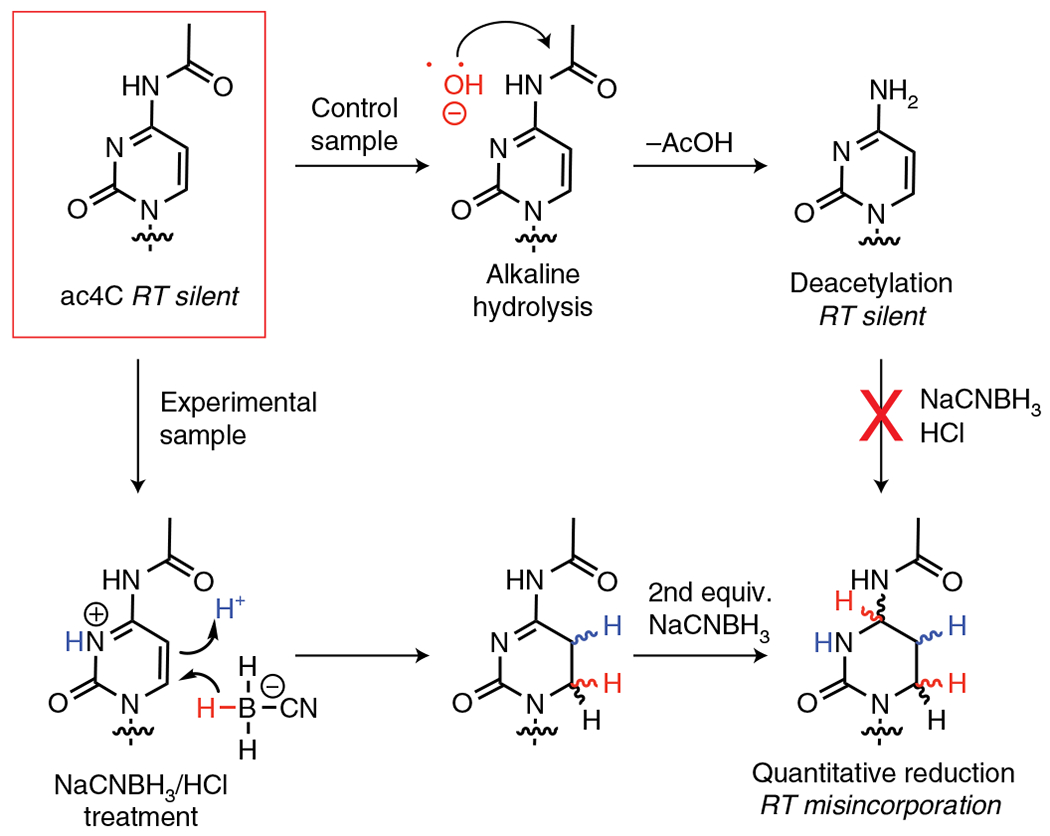
Endogenous ac4C forms base pairs identical to cytidine, rendering it silent in RT and cDNA sequencing experiments (top left). Treatment of ac4C-containing RNA with NaCNBH3 under acidic conditions results in formation of a reduced nucleobase, which is read as a ‘U’ during RT and results in misincorporations that can be detected by cDNA sequencing (bottom right). This chemistry is blocked when ac4C is chemically deacetylated by alkali hydrolysis (top right), which provides a control for specific detection of the acetylated nucleobase. equiv., equivalent.
