Fig. 2 |. Experimental workflow for ac4C-seq protocol.
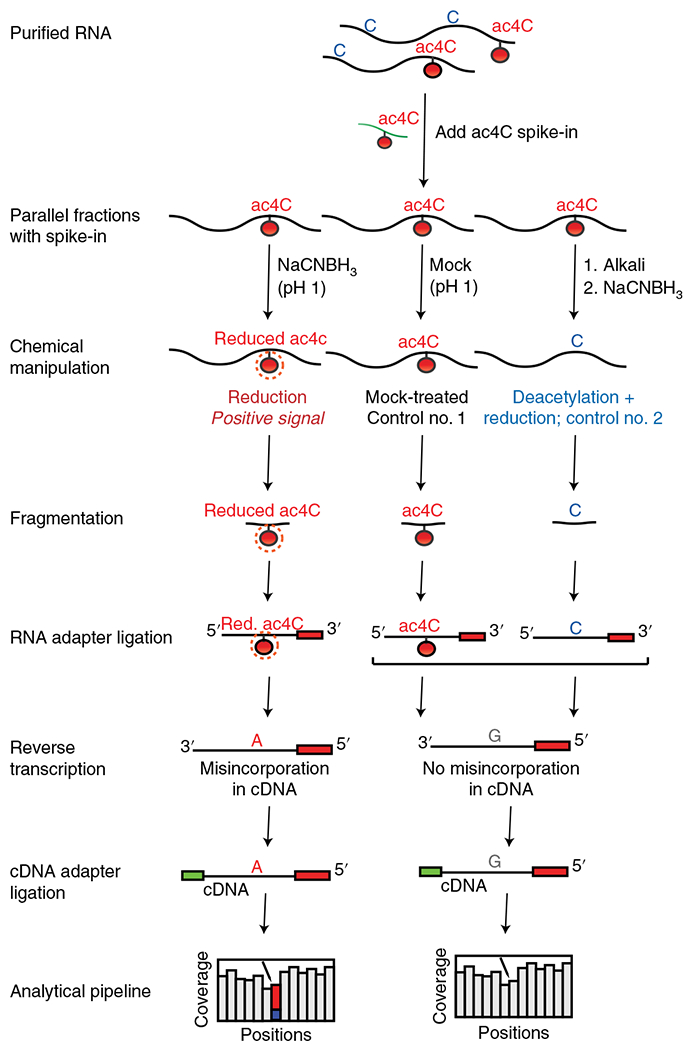
RNA is spiked with a synthetic ac4C RNA spike-in and split into three samples. One experimental sample is treated with NaCNBH3 under acidic conditions (reduction, positive signal), while two control samples are subjected to acidic conditions without reducing agent (mock-treated, control no. 1) and deacetylation followed by NaCNBH3 treatment (deacetylated + reduction, control no. 2). A 3′ adapter is ligated onto fragmented RNA, which is then reverse transcribed, resulting in misincorporation of ‘A’ in cDNA at positions of reduced ac4C. A 3′ adaptor is ligated to facilitate cDNA library construction, with subsequent sequencing and bioinformatic analysis being used to identify ac4C-modified sites. Figure adapted with permission from ref. 5. Red., reduced.
