Fig. 3 |. Flowchart illustrating steps involved in ac4C-seq.
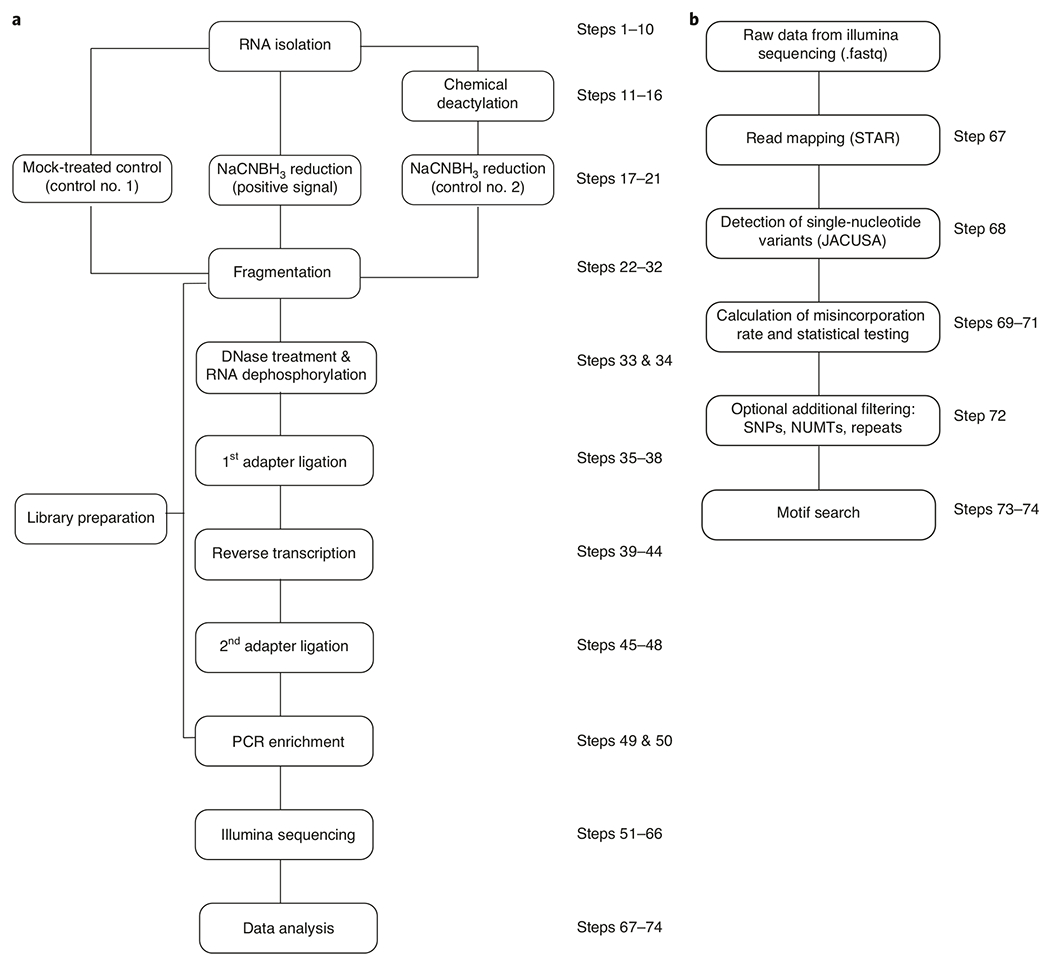
a, First, RNA is isolated, followed by chemical treatment to prepare experimental and control samples. These RNA samples are used to prepare cDNA libraries that undergo next-generation sequencing and are subsequently analyzed by using computational pipelines. b, Data analysis includes mapping of sequenced reads to the appropriate genome, calculating misincorporation rate in treatment and control samples and conducting statistical tests to identify ac4C sites. After additional filtering, motif analysis is applied to strengthen confidence in newly identified candidate ac4C-modified sites. NUMT, nuclear mitochondrial DNA; SNP, single-nucleotide polymorphism.
