Abstract
Tissue nanotransfection (TNT) is an electromotive gene transfer technology that was developed to achieve tissue reprogramming in vivo. This protocol describes how to fabricate the required hardware, commonly referred to as a TNT chip, and use it for in vivo TNT. Silicon hollow-needle arrays for TNT applications are fabricated in a standardized and reproducible way. In <1 s, these silicon hollow-needle arrays can be used to deliver plasmids to a predetermined specific depth in murine skin in response to pulsed nanoporation. Tissue nanotransfection eliminates the need to use viral vectors, minimizing the risk of genomic integration or cell transformation. The TNT chip fabrication process typically takes 5–6 d, and in vivo TNT takes 30 min. This protocol does not require specific expertise beyond a clean room equipped for basic nanofabrication processes.
Introduction
Tissue reprogramming can be used to repair damaged or diseased tissue and may enable development of patient-specific therapies where a part of the patient’s own body is utilized as a bioreactor1,2. However, achieving safe and efficient in vivo transfection and tissue reprogramming remains a challenge. We developed tissue nanotransfection (TNT), a non-viral approach to topically deliver reprogramming factors through a nanochanneled device in vivo. In the TNT approach, a focused electric field is applied to a tissue through arrayed hollow needles, to locally nanoporate cell membranes and deliver biomolecules directly into cells in vivo without the need for elaborate laboratory infrastructure or viral vectors3.
In this protocol, we describe how to fabricate a chip containing silicon hollow needles of variable shapes and adjustable bore diameter that is used alongside electroporation for in vivo tissue reprogramming. We have previously used this protocol to reprogram mouse tissue in vivo3. The fabrication process is intricate; however, in this protocol, we include full details to facilitate reuse. By following this protocol, nanoscientists should be able to independently reproduce the fabrication of the TNT chip for any application of their choice. We also include approaches to address potential problems and troubleshoot issues at all stages of the protocol.
Development of needle-based gene-delivery strategies
Gene-delivery platforms for therapeutic purposes need to ensure that delivery is reproducible and appropriately localized4. Currently, two types of delivery methods are commonly used. In one approach, a cell membrane–permeabilization-based method involving mechanical5,6, thermal7,8 or electrical means9,10 may be used to enhance cell membrane permeabilization to enable direct delivery of cargo into the cytosol. Alternatively, cargo may be encapsulated in carriers such as viruses11,12, exosomes13,14, vesicles15,16 or nanoparticle capsules17, which can be readily transported into the cell through endocytosis uptake mechanisms or membrane fusion. Among membrane permeabilization–based methods for delivery, bulk electroporation (BEP) is widely used because of its high transfection efficiency, even for hard-to-transfect cell types such as T-cells18. However, BEP uses a strong electric field that is often associated with damage to the cell membrane18. Our motivation to develop an alternative to BEP stemmed from the observation that most cells survive after BEP but with reduced plasticity that prohibits further reprogramming.
In general, efforts to miniaturize BEP to the micro- and nanoscale have enhanced both efficacy and safety2. Such miniaturization allows for the use of high-intensity electric fields that locally nanoporate cell membranes, enabling delivery of a desired cargo, such as nucleic acids, into the cell cytoplasm through iontophoresis with reduced cytotoxicity compared to BEP2. An alternative approach, nanoelectroporation, uses nanostructures to induce localized electroporation, which allows for the effective use of a low-voltage electric field, thus minimizing cell damage18. As opposed to BEP, nanoelectroporation induces localized and well-defined pores on the cell membrane for gene transfer to enable cell and tissue reprograming2,3,9,18–20. To minimize BEP-induced cell damage, we developed a nanoscale transfection technology for in vivo tissue reprogramming3. We designed a silicon wafer patterned with an array of 10-μm-deep, ∼500-nm-wide nanochannels within larger reservoirs that can be topically applied to tissue in vivo to achieve high delivery efficiency3. The hollow-needle array structure of the TNT chip enables efficient cutaneous delivery of loaded cargo21,22.
Development of silicon hollow-needle arrays
Several techniques are available to fabricate different types of hollow structures, including laser cutting23, lithographically patterned molding24, X-ray exposure25, laser-initiated polymerization26, pyrolysis of polymer precursor27 and wet chemical etching combined with deep reactive ion etching (DRIE)28,29. Silicon is the preferred choice of material and is widely adopted for miniaturized needle array fabrication because of its excellent mechanical properties30,31, biocompatibility for retaining cell viability32,33 and compatibility with conventional semiconductor processes. Wet chemical etching has also been used to fabricate solid-type silicon needles, using a different etch rate between crystal planes in an alkaline solution such as KOH31. The limit in anisotropy can be removed by combining wet chemical etching with an isotropic dry etching process30,31. The two-step Bosch process uses sequential cycles composed of passivation and etching steps switched on and off by fluorine-based chemistries to etch silicon. The Bosch process can be used to fabricate long needles30,33–35; however, the etching rate decreases rapidly as the aspect ratio increases and finally reaches a constant low value33,36,37, which limits the fabrication of high-aspect-ratio hollow structures.
Although silicon-based needle arrays are a powerful tool for various drug-delivery applications and are compatible with prevailing semiconductor fabrication processes, there remain technical difficulties for obtaining silicon hollow-needle devices with a high aspect ratio. To overcome the challenge of aspect ratio–dependent etching, we adopted the three-step Bosch process composed of passivation, clearing (to remove passivated polymers using active argon) and etching steps. The detailed fabrication of silicon hollow needles using the three-step Bosch process is described in Step 1A(xxiii).
Applications
Silicon hollow needles provide excellent drug-delivery performance when combined with electroporation technology. When combined with transdermal electroporation, silicon hollow needles enhance cell permeability, enabling efficient drug delivery35.
TNT chips with silicon hollow-needle arrays were designed primarily to achieve in vivo gene delivery via electroporation, with the end goal of reprogramming tissue. We have previously demonstrated that non-viral topical TNT can achieve in vivo reprogramming of tissue in a murine model1–3,38. TNT initiated the conversion of skin cells to either functional blood vessels or to electrophysiologically active neurons in vivo3. Vasculogenic TNT-based skin reprograming prevented development of tissue necrosis in a severe mouse model of hindlimb ischemia3. Neurogenic skin reprogramming produced electrophysiologically active neural cells, which were transplanted into the stroke-affected site of mouse brains and improved recovery3.
It is important to follow the fate and function of reprogrammed cells in vivo after cell and tissue reprogramming to better understand the process3. Our recent work revealed additional insight into the bystander effects of the process, where therapeutic benefit is derived from secreted cues of treated tissue, rather than the treatment itself. We found that under conditions of neurogenic reprogramming, cells in the skin that did not convert into neural cells but were in close proximity to cells that had switched to a neurogenic fate produced copious amounts of neurotrophic factors20. This neurotrophic enrichment of the affected skin tissue enabled the reprogrammed cells to develop over a month, resulting in mature functional neural cells. The neural cells generated in the skin were not part of any neural circuit at the site of reprogramming. However, when transplanted to the stroke-affected brain, the cells were able to improve recovery from acute ischemic stroke3. Delivery of neurogenic reprogramming factors may also result in neurotrophic enrichment of the skin tissue matrix, which can be leveraged to preserve pre-existing nerve fibers in diabetic skin that may otherwise degenerate20. After TNT of mouse skin, transfected cells produced exosomes loaded with the delivered reprogramming factors, which may enable more widespread cargo delivery and further downstream therapeutic benefits3.
Although there are currently a few published examples of TNT for murine skin applications3,20,39, we hypothesize that TNT could be applied to various tissues in the living body40. Because TNT is capable of achieving tissue reprogramming in immunocompetent in vivo settings, the reprogrammed tissue is likely to be stable and well tolerated by the body.
We also anticipate that minor adjustments to optimize our TNT platform could enable this method to be adapted for compliance with appropriate regulatory agencies for human and clinical applications. A broad range of applications such as gene editing, transdermal drug delivery, nano-carrier delivery and biomarker sampling from skin would thus be feasible using the TNT chip described in this protocol.
Comparison with other methods
Alternative methods of in vivo reprogramming involve transfecting cells in vitro followed by implantation41–43. Transfecting cells in vitro involves various regulatory and laboratory infrastructure hurdles, and cell implants often have low survival and poor tissue integration. Common methods for bulk in vivo transfection include delivery of viral vectors or electroporation, both of which can lead to undesired immune reactions44. Many viral vectors cause long-term expression of the transduced gene, which can result in insertional mutagenesis and genomic integration12,45. Although long-term gene expression is useful for some applications of gene therapy, for applications in which sustained gene expression is unnecessary or even undesired, transient transfection must be ensured. Electroporation-based methods have been reported to enhance the efficiency of plasmid penetration in the target cell or tissue; however, these methods are often limited by heterogeneous delivery46–48. In contrast, TNT achieves localized non-viral gene delivery with direct conversion of tissue function in vivo without the need for any complex laboratory procedures or ex vivo cell manipulation. In addition, TNT delivers reprogramming factors in a relatively noninvasive way in a short time (1.1 s for the example described here). Delivery of plasmids by using TNT enables temporal and spatial control of the expression of multiple genes3,20,39.
Silicon hollow-needle arrays have advantages compared to hollow-needle arrays fabricated from other materials. Development of metal microneedles is limited by challenges in micromanufacturing capabilities and high production cost. However, metal microneedle structures are able to easily penetrate the skin and effectively deliver drugs to tissue as a consequence of their mechanical and electromechanical properties25,49. Different polymer-based needle arrays have been studied owing to their excellent biodegradability, biocompatibility and capacity to be produced at large scales at low cost24,50–54. However, some disadvantages of polymer-based needles are their low strength and toughness compared to other materials such as metals and silicon. Among the available fabrication materials, silicon is the most widely used because of well-established silicon complementary metal–oxide–semiconductor and micro electromechanical systems technologies that allow wafer-scale production of desirable shapes and sizes of silicon needles28,34,35,55,56. Silicon needles also have reasonable mechanical strength (sustaining a pressure of ∼0.65 MPa; see Supplementary Manual) for biointerfacing, and trace silicon (if needles penetrate the skin) can be eliminated from the body within several weeks without biotoxicity21,32. Large-scale production capabilities, standard fabrication processes and high biocompatibility make silicon hollow needles suitable to be integrated in transdermal electroporation systems for use in TNT technology.
There are several alternative approaches to TNT that can be used to deliver reprogramming factors. Nanoneedle- or nanostraw-based electroporation systems have also been reported for in vitro delivery of reprogramming factors; however, limited in vivo applications have been described57–62. Metallic, polymeric, silicon and other inorganic needles23,24,26,51,53,55,63–65 or nanoneedles66–68 have also been reported to deliver cargo molecules in the absence of electroporation. Dissolvable polymeric needles can also be buried in the target tissues for controllable delivery over specific time periods51,53. These passive delivery methods rely on the diffusion of cargo through physiological environments, and the delivered molecules may be taken up into cells by endocytosis or membrane fusion69,70. In contrast, electroporation-based delivery methods provide electrophoresis-assisted delivery through transient cell membrane pore formation to shuttle cargo directly into the cytosol.
Limitations
Limitations of electroporation include risk of tissue injury and operational complexities, including the need for specific instruments and predefined electrical parameters for the electroporation process. Safety concerns can be addressed by optimizing electric pulse voltage to maintain cell viability71–73. We acknowledge that the fabrication process and needle array–based TNT technique could be further improved. Several steps in the TNT fabrication process are not compatible with standard semi-conductor processes and therefore would limit scalability. For example, in Step 1A(xxii) of the TNT fabrication process, a polyimide film tape with silicon adhesive is used to cover the backside hollow channels to avoid gas leaking in the following etch (Step 1A(xxiii)), which is a simple and effective solution for laboratory-based fabrication that would need to be further adapted for increasing scale.
Although this protocol can generate high-aspect-ratio silicon hollow-needle arrays, the tapered geometries and rough walls within the hollow bore may cause turbulence or adversely influence the flow behavior of the liquid cargo, depending on the level of roughness and the average velocity of the liquid. Although attention to these potential limitations is recommended, these factors are not likely to complicate the intended application. The roughness mainly exists at the connection part between the upper narrow core (∼9 μm in diameter) and lower wide core (∼25 μm in diameter) caused by the imperfect DRIE process due to difficulties in cleaning the residual passivation polymers within the high-aspect-ratio hollow structure. The relative roughness (ratio of roughness and diameter) is estimated to be <0.08. Such roughness is likely to decrease the flow rate by a ratio of ∼0.3 according to previously reported experiments74. This roughness could be reduced by further optimizing the DRIE process75,76. The influence of roughness can be minimized by adjusting the applied voltage and pulse duration to improve delivery. To obtain the deeply etched structure with a highly straight wall, one must overcome the ratio-dependent etching effect by more advanced techniques, such as by modifying the physical parameters or by further adjusting the etching parameters in the Bosch process (Supplementary Manual)29.
Although the silicon needle shows excellent strength under compression forces (Supplementary Manual), longer needles can be damaged by strong bending or shear force30,31,35. To avoid such damage, the development of standardized procedures is necessary not only for the fabrication process but also for the application of TNT to target tissues. The device should be stored in appropriate housing for protection, such as an airtight plastic container at room temperature (15–30 °C). Loading the device vertically on the target tissue is important to avoid bending or shear force. Operators must retract the needles vertically after finishing the TNT treatment. To overcome the limitation in brittleness, there are also alternative approaches to fabricate hollow needles with strong mechanical properties, such as fabrication of needles with metal/semiconductor/oxide deposition layers on predefined polymeric molds; however, these alternative materials may be limited in terms of biocompatibility24,55.
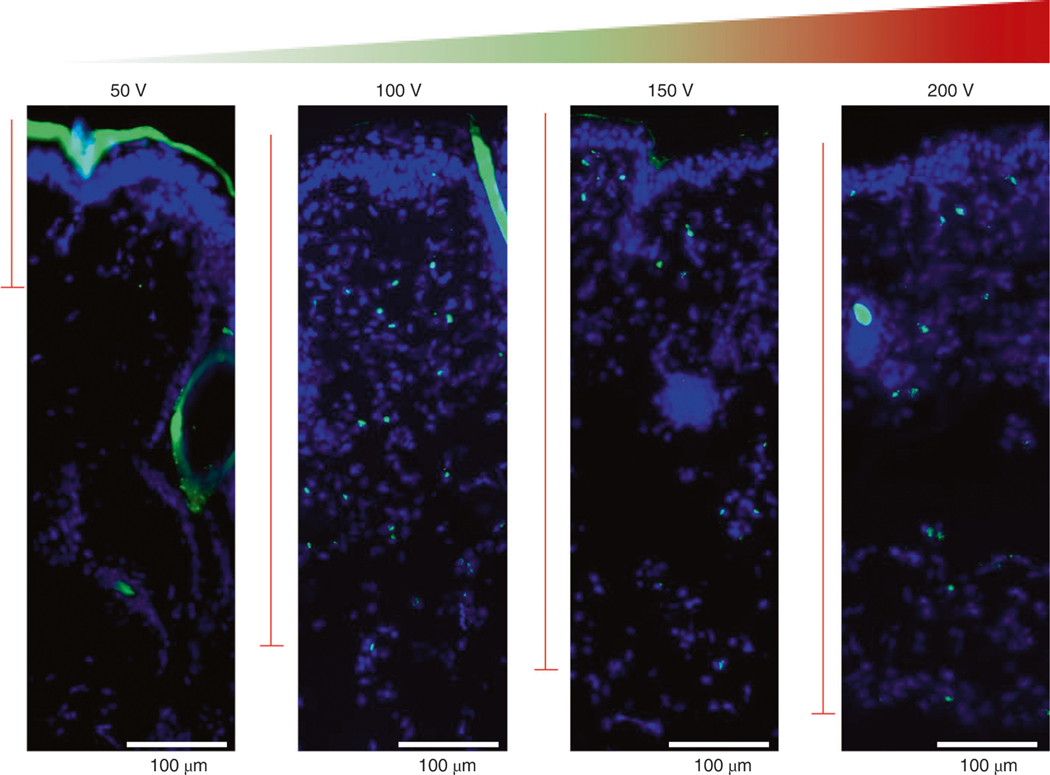
The dimensions of the fabricated chip limit three aspects of needle array–based TNT: treatment area, contact situation and insertion depth. As a result, concerns regarding effectiveness, feasibility and safety may arise in complex situations where more intimate contact, longer delivery distance and lower operation voltage are required. For surface treatments, TNT-mediated delivery requires exfoliated skin tissue to ensure sufficient contact between the target skin tissue and the surface of the chip. Likewise, internal organs are covered by membranes or fascia that need to be bypassed to transfect the organ. Curvature and roughness of skin can result in air gaps between the point of contact and the chip, which may lower efficiency of drug delivery and reduce the uniformity of drug distribution, because the chip is designed for optimal use on a flat surface. Using needles of different sizes and heights on the TNT chip allows for some control over the penetration depth for cargo delivery, and additional control in depth of penetration can be gained by modulating the strength of the applied electric voltage. Within a narrow range (50–250 V), higher voltages increase the depth of penetration.
Each chip can typically be reused 10–25 times (depending on tissue type) if handled carefully and thoroughly cleaned after use. Some process steps such as backside taping (Step 1A(xxii)), DRIE (Step 1A(xxiii)), dicing (Step 1A(xxvii)) and wet cleaning (Step 1A(xxviii)) can cause clogging of the hollow channels of the needle. Repeated use of the TNT chip may also result in clogging of the hollow channels. Clogging is more likely when the target tissue is not thoroughly cleaned (e.g., insufficient exfoliation of superficial dead cells on the skin). Thus, cleaning and chip replacement are probably necessary.
This protocol describes electroporation-mediated topical transfection of plasmids to mouse skin with the goal of transient gene expression to facilitate tissue reprogramming. Correct insertion of the positive electrode into the skin is an important preparatory step for electroporation. The insertion position and depth must be optimized for each application. This helps standardize the distribution of the electric field intensity, resulting in uniform delivery. Use of excessive voltage during electroporation may cause tissue inury72,77,78. At high electric field strengths (e.g., >200 V/mm), the irreversible pores in cell membranes and changes in physiological activities can compromise tissue or cell viability73. Numerous strategies have been developed to lower the applied voltages used during electroporation, including the combination of microfluidic devices79,80 and the use of membranes containing nanopores3,19,81 or nanostraws57,59. The needle arrays we describe here reduce the need for high voltages because they optimize contact with the tissue.
Overview of the procedure
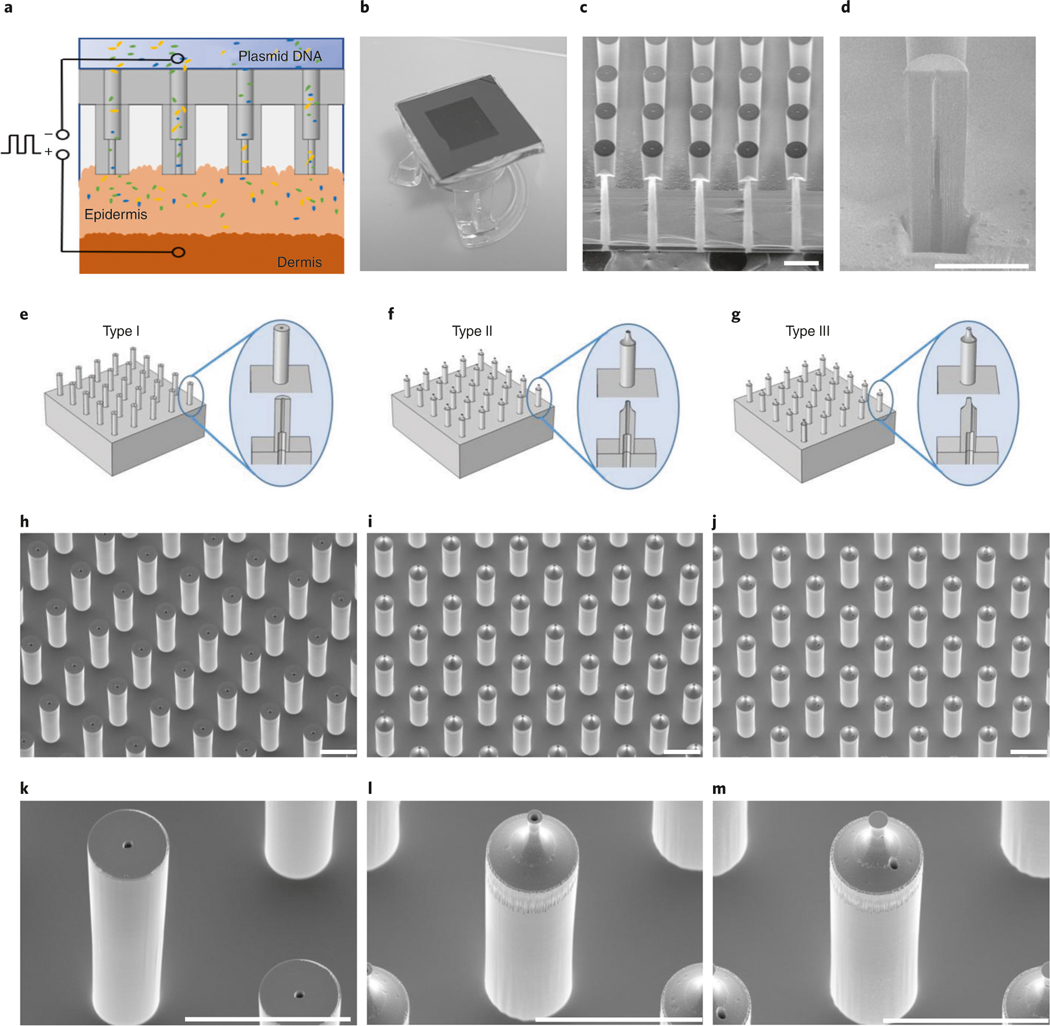
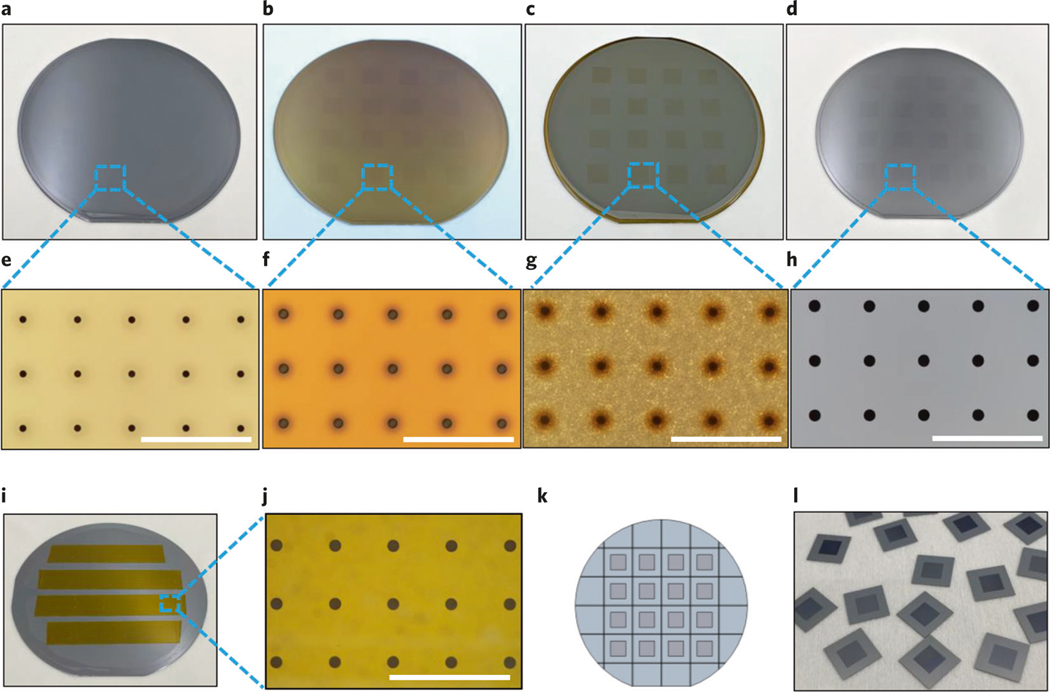
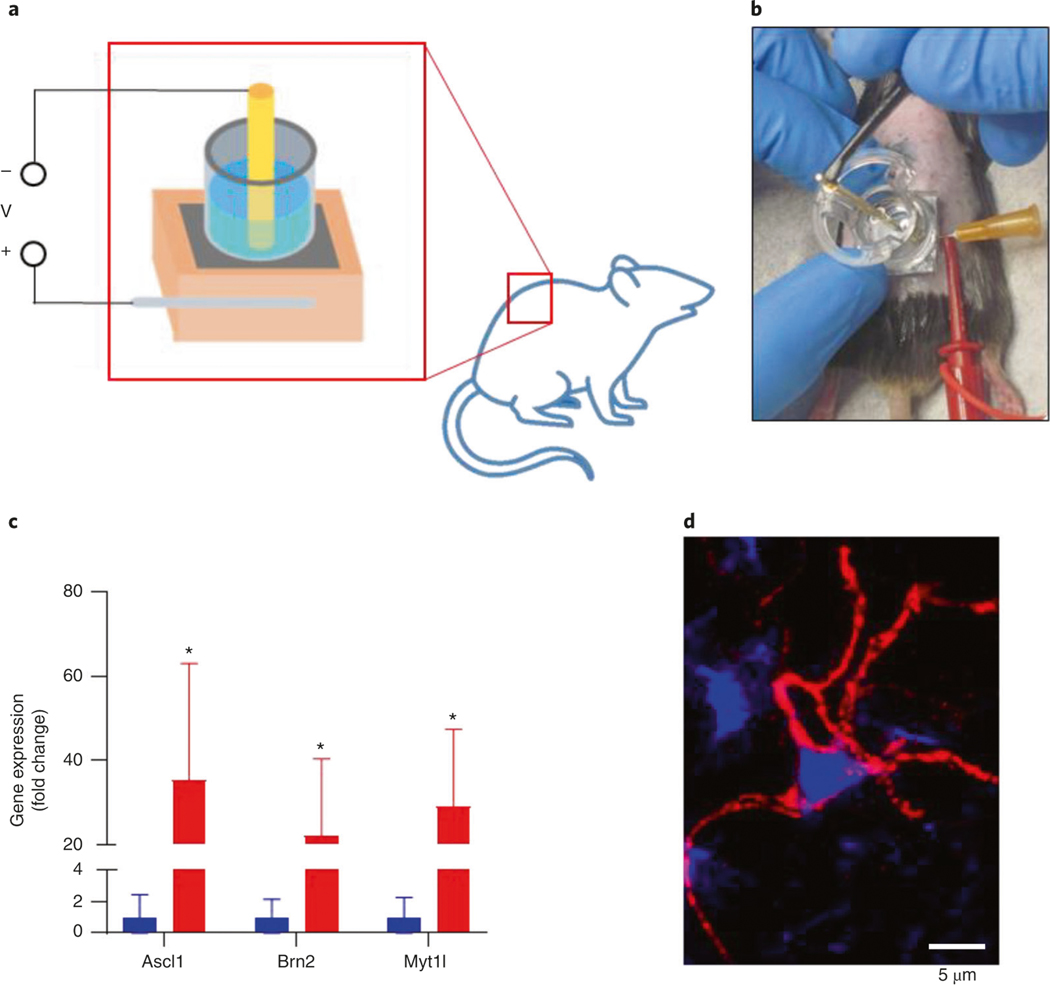
The overall TNT process involves three parts: fabrication of the silicon hollow-needle array, preparation of the TNT device and in vivo TNT application. Before starting the procedure, it is important to consider the optimal array design for the proposed application. For topical skin delivery applications, we recommend using a silicon hollow-needle array with flat tips (Step 1A). For deep tissue delivery applications, we recommend using a silicon hollow-needle array with sharp tips to facilitate tissue penetration (Step 1B or 1C). Figure 1a is a schematic of the TNT chip used for delivery of reprogramming factors into skin tissue assisted by transdermal electroporation. Figure 1b shows a photograph of a fabricated silicon hollow-needle array mounted on a transwell insert that serves as a cargo reservoir.
Fig. 1 |. A schematic view of the TNT process, chip designs and scanning electron microscope (SEM) images of the fabricated chips.
a, Schematic diagram of the TNT process conducted on exfoliated skin mediated by hollow needles. The plasmid DNA can be delivered to the skin tissue under a millisecond square electric pulse. b, A typical TNT chip with needle array mounted on the reservoir of plasmid DNA solution. c, A typical SEM image of the fabricated primary Type I needles showing the bores buried in the silicon substrate, d, An SEM image of an opened bore within a single needle, taken at a tilted angle of 52°. The needle is measured with an ∼9-μm inner diameter for the upper lumen, an ∼25-μm diameter for the lower lumen and an ∼65-μm outer diameter for the needle after PECVD SiO2 coating. The narrowest inner diameter of the upper lumen is ∼4 μm. The height of the needle is ∼210 μm. e, Design of the primary Type I hollow-needle array with flat tip. f, Design of the type II hollow-needle array with sharp tip and centered bore. g, Additional Type III hollow-needle array with sharp tip and off-centered bore. h, A typical SEM image of the fabricated primary Type I silicon hollow-needle array with flat tip. i, An SEM image of the fabricated Type II silicon hollow-needle array with sharp tip and centered bore. j, Additional Type III silicon hollow-needle array with sharp tip and off-center bore. k–m, The zoomed-in views of needles in h–j, respectively. Images were taken at a tilted angle of 30°. Scale bars, 100 μm.
Experimental design
Fabrication of silicon hollow-needle arrays
The fabrication process begins with a double-side polished 4-inch silicon (Si 100) wafer topped with a layer of photoresist, through which the desired pattern is lithographically defined and developed for DRIE using the Bosch process. By modifying the lithography mask and etching process, this protocol describes how to produce a primary needle array with center bore and flat tip (Fig. 1e, Type 1). Two additional examples of sharp tips (Type II) or sharp tips with off-center bore (Type III) are also described in detail (Fig. 1f,g). The latter two types of needle arrays with sharp tips are probably useful in reducing insertion force to easily penetrate into tissue. Shrinking the hollow diameter of the needle will result in an enhanced local electric field and an increased velocity of the cargo at the outlet of each needle; however, it may also limit the total amount of sample flow through the needles. Clogging is generally observed in hollow needles with small diameter (<5 μm). This issue can be minimized by optimizing the bore diameter and by using an off-center bore structure (Type III). The bore diameter of the needle can be adjusted to a target value (dependent on tissue type and depth of delivery required) by using plasma-enhanced chemical vapor deposition (PECVD) to deposit a conformal oxide coating. In addition, a transwell insert is attached to each silicon chip to serve as a reservoir for plasmid DNA cargo.
Although cleanroom-mediated semiconductor fabrication techniques are familiar to researchers, to obtain high-quality fabrication results for the needle arrays, two specific aspects of this procedure require special attention for successful TNT production. The first is precise alignment of the patterns when flipping the wafer to fabricate the hollow channels on one side (Step 1A(xix)), and the second is gentle handling to avoid clogging phenomena in the hollow bore of the needles (Step 1A(xxii, xxvii and xxiv).
The critical parameters of needle design that affect TNT outcome are the final nozzle size (smallest hollow bore size after PECVD coating), needle length and density. Parameters such as needle outer diameter, original inner diameter and the diameter of the reservoir hole are not crucial for gene delivery but offer flexibility in the design of TNT.
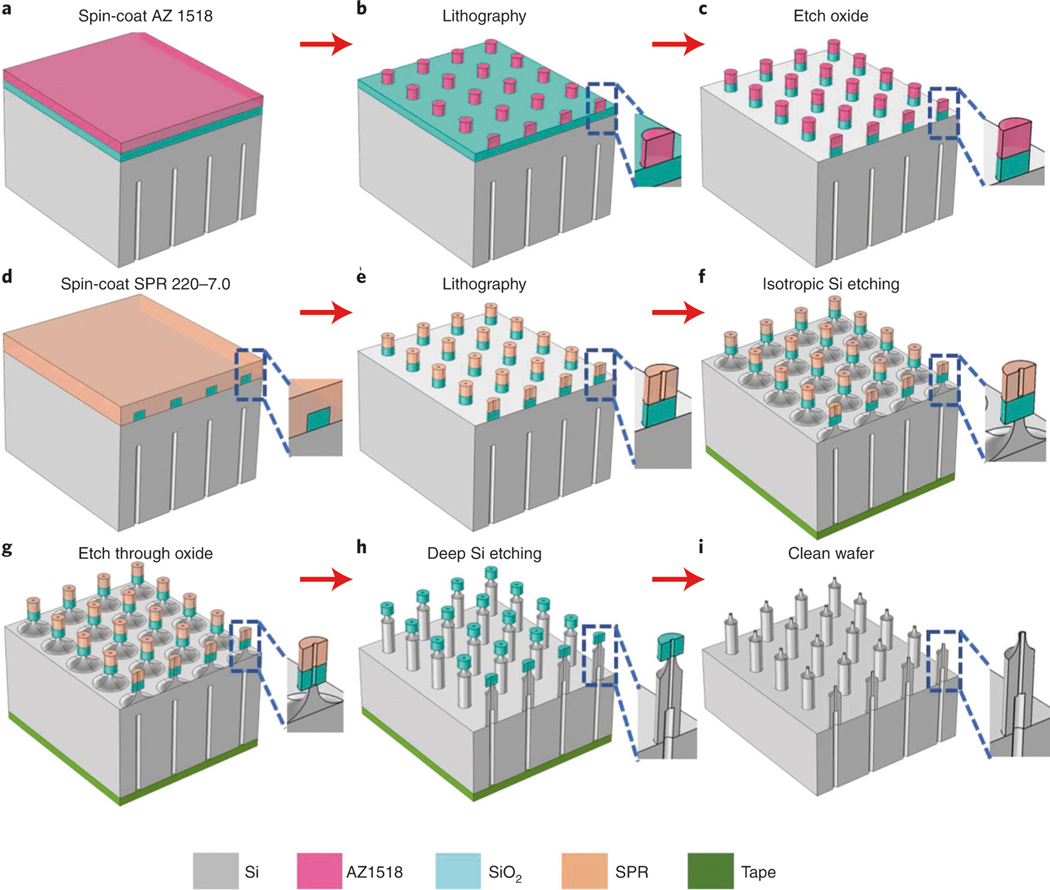
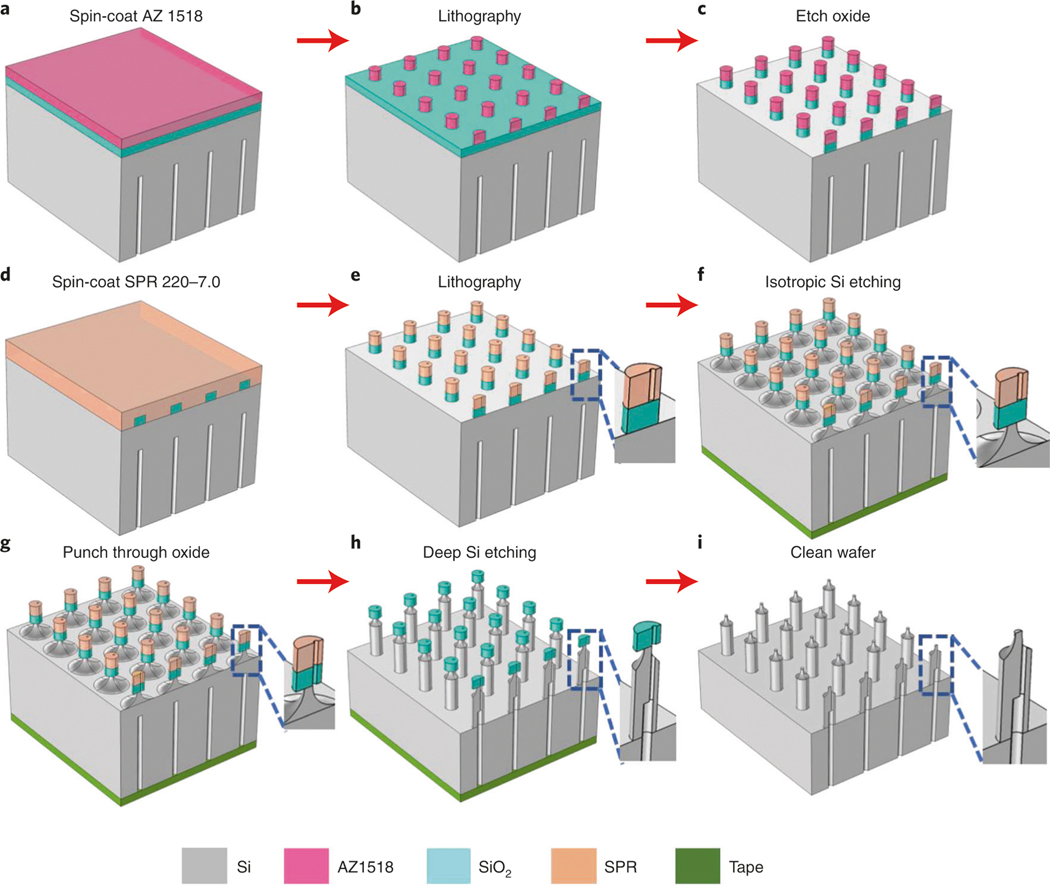
Fabrication of silicon hollow-needle arrays with flat tip (Type I)
The silicon hollow-needle array was developed using a semiconductor process that should enable reproducible fabrication of chips at large scale to lower future fabrication costs. We used semiconductor fabrication processes, including optical lithography and dry etching, to optimize the needle types, inner structures, materials and functionalities. A detailed fabrication flow of the Type I needle arrays is described in Step 1A(i–xxix). It is worth pointing out that to achieve the high-aspect-ratio hollow-needle array, the three-step Bosch process is used in this protocol rather than the traditional two-step process. The improved Bosch process for high-aspect-ratio structures is composed of passivation, clearing (removal of passivating polymers) and etching steps with fast gas switching to achieve a vertical and smooth structure. Instead of the commonly used oxygen, active argon is used to efficiently remove the passivated polymers on the top surface in the clearing step. This is a crucial step to mitigate the accumulation of undesired chemistries and irregular shape on the wall of the hollow structure. Finally, a layer of SiO2 is deposited on the fabricated needles to tune the bore size.
Alternative designs of silicon hollow-needle arrays
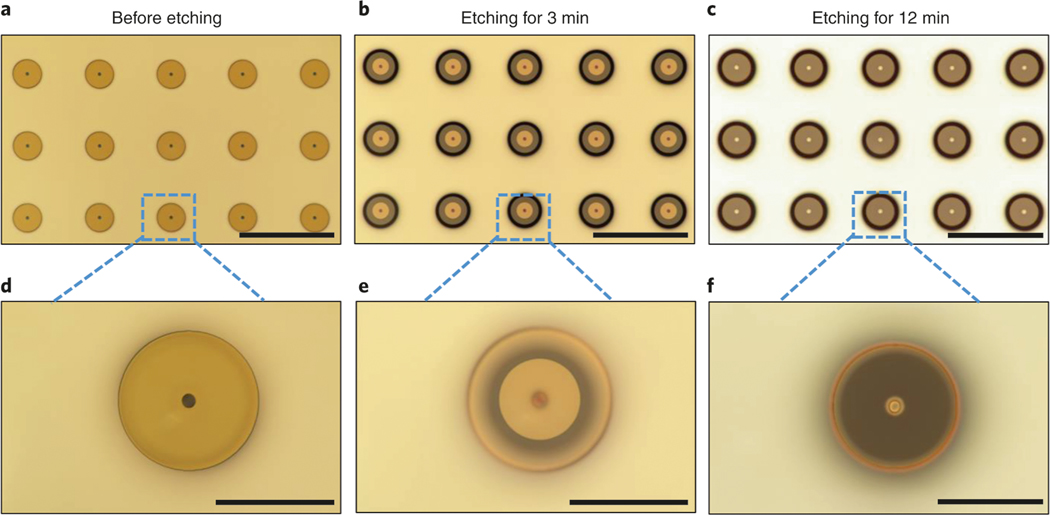
To decrease the insertion force for penetrating skin or muscle more easily, needle arrays with sharpened tips were developed with hollow bores on-center for Type II (Step 1B(i–xx)) and off-center for Type III (Step 1C (i and ii) structures. The fabrication process of the sharp-tipped hollow-needle array follows similar steps as the flat case, using both lithography and the Bosch process. However, the critical step is that the sharp structure is generated by isotropic dry etching around the tip of the needles. Delicate control of the etching time enables the needle tip to be defined with the desired sharpness with a diameter as small as ∼10 μm. Too short or too long of an etching time will cause under- or over-etching, respectively, resulting in non-uniform and undesired shapes. Furthermore, by modifying the process, we fabricated sharp-tipped hollow needles with an off-center bore (Type III). Compared with Type I and Type II, this alteration in process enables a shift of the hollow bore of the needle with a deviation of 15 μm from the center of a 55-μm-diameter needle.
The tip profile can be improved by optimizing the etch bias82 and/or oxygen content83 in the SF6based dry etching system. It can also be improved by KOH-based wet etching34,84. Increasing the tip sharpness is a simple way to reduce the resistance during the insertion process. The Type II needle can effectively create pathways on the target tissue through micron-scale tip sharpness. However, a proper needle density, in addition to tip sharpness, is also required to reduce the resistance and improve the efficiency of creating pathways for cargo delivery into tissue85.
Quality control in the fabrication process of the needle array
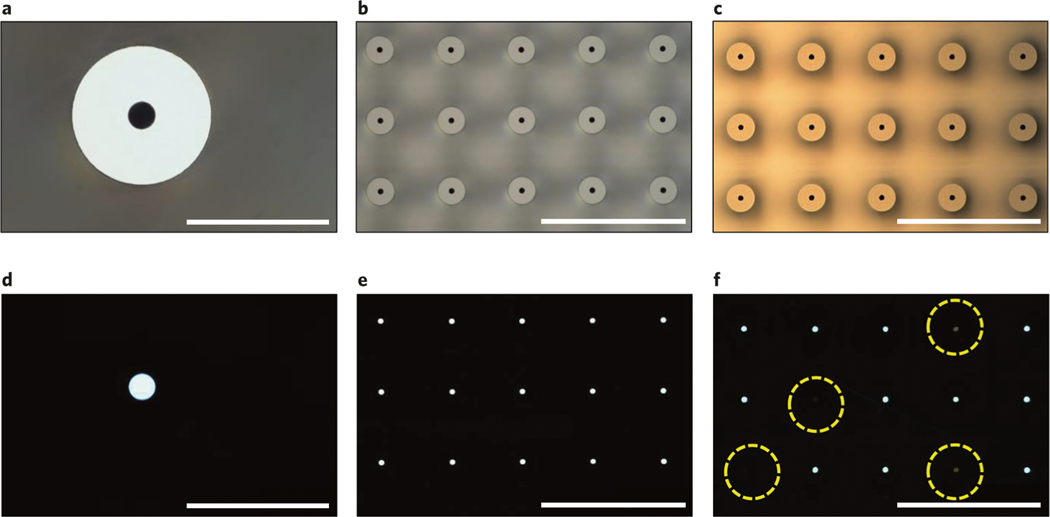
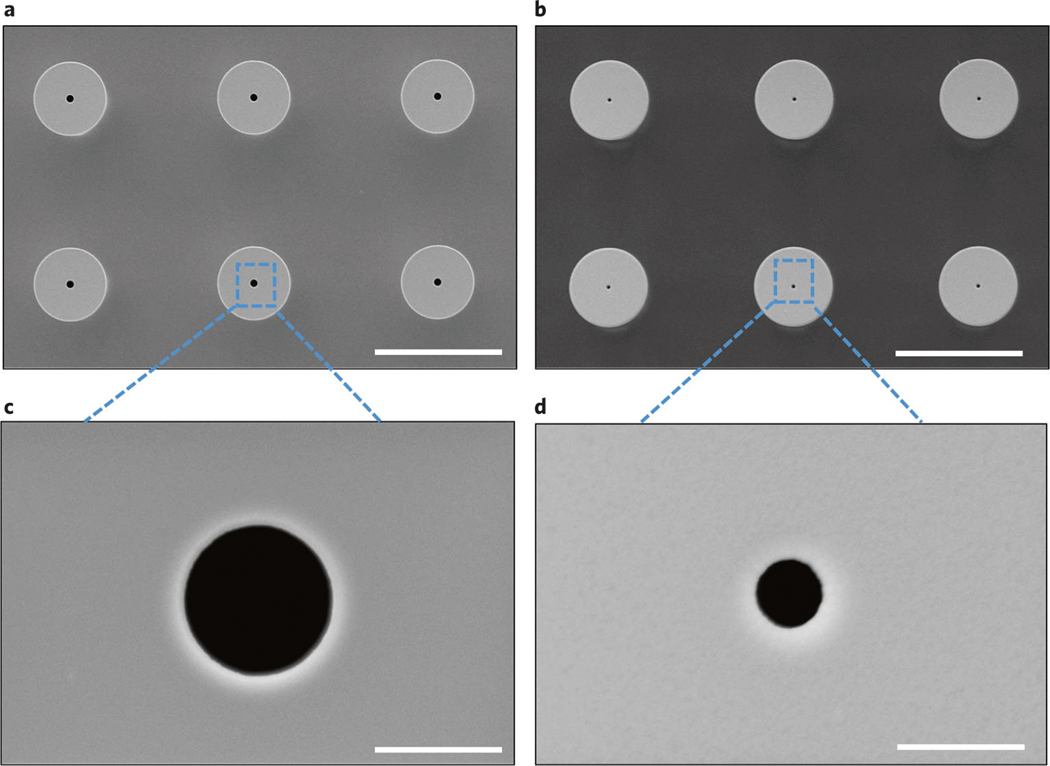
The fabrication process of hollow-needle arrays is vulnerable to three types of defects, including clogging, misalignment and damage. In the hollow-needle array structure, it is critical that the channels be fully developed in the etching process and clear of particles so that drugs can be successfully delivered through the lumen. Optical transmission microscopy can be used to check the quality of each TNT chip and investigate if there are any blockages in the channels. Figure 2a–c present optical images of the TNT chip showing the hollow-needle arrays, while their transmission images are shown in Fig. 2d–f, respectively. The white light uniformly transmits from each hollow microchannel at the center of the needle, which is evidence of a clear hollow channel, ensuring that the liquid cargo can freely flow through it. In Fig. 2f, some of the hollow channels exhibit clogging, caused by either an imperfect etching process or the existence of undesired particles from backside taping (Step 1A(xxii)), DRIE (Step 1A(xxiii)), dicing (Step 1A(xxvii)) and cleaning (Step 1A(xxviii)). Another issue is that misalignment may arise in this fabrication process, because etching steps are separately performed on two sides of the wafer. It is necessary to carefully align the predefined markers (Step 1A(v)). If there is misalignment between the upper hollow bores and the lower counterparts of the needle arrays, it can be observed by looking at the cross-section of the array. Some needles may be damaged during the fabrication process, caused by unexpected mechanical shocks in operation steps such as ultrasonic cleaning and mishandling; hence, special care is warranted. To avoid these potential problems during the fabrication process, each step must be handled with caution and precision as detailed below. Totally avoiding all the imperfections is hard; therefore, threshold values are set for an individual device to be acceptable for use. The individual device will be accepted if the measured mean deviations of geometrical parameters are within the errors shown in the corresponding steps of the fabrication process. An individual device is considered inadequate for use if >10% of the needles are clogged or misaligned.
Fig. 2 |. Quality control: clogging problem in the fabricated hollow needles.
a, A typical optical microscopy image of a needle. b, An optical image of a needle array where no needles are clogged. c, An optical image of a needle array where some needles are clogged. The transmission optical microscopy images, a–c, are correspondingly shown in d–f, respectively. Four hollow needles show a clogging effect, highlighted by the yellow dashed circles in f. Images were taken from the top view. Scale bars, a and d, 60 μm; b,c,e and f, 300 μm.
Preparation of the TNT device
After the chips are fabricated, preparation steps are required to integrate the fabricated chip and the electroporation apparatus to enable gene delivery (Steps 2–8). The simplest integration of this system consists of a TNT chip mounted with a reservoir to hold an aqueous solution containing the cargo. In addition, an electric pulse generator with precision control is necessary to achieve nanoelectroporation. Polydimethysiloxane (PDMS) is used to mount the reservoir on the TNT chip (Steps 2–8). PDMS is mixed with hardener to form viscosities with different weight ratios, which can be used to make PDMS films with an embedded funnel attached to the chip. Oxygen plasma treatment is used to bind the chip to the PDMS. Here, both the top side of the chip and the bottom side of the PDMS film have been treated using a plasma oven and then gently pressed together, providing a complete bonding between layers. The thickness of the PDMS films has been adjusted to be 2–4 mm. Thicknesses higher than 4 mm are not flexible enough to provide complete leak-proof bonding between the chip and could also damage the chip in the attachment process. Thinner films (<2 mm) are too flexible and thus generate wrinkles and air gaps between the chip and the film.
Quality control in the preparation of the TNT device
The TNT device after mounting of the reservoir is mainly vulnerable to two types of defects: damage and leakage. The assessment of damage can be checked by transmission microscopy. To test for any leakage, the TNT device is placed on a Whatman filter paper, and the reservoir is filled with 0.01% (wt/vol) bromophenol blue solution and incubated for 2–5 min. Any leakage from the device will turn the filter paper blue, in which case the device needs to be remounted with the reservoir again to ensure proper sealing between the reservoir and the chip.
Plasmids
Although we anticipate that it should be possible to deliver any plasmid of interest, here we provide an example of delivery of three plasmids: Ascl1, Brn2 and Mytl1 gene were cloned with pCAGGs plasmid backbone and constructed commercially by ABM Good (www.abmgood.com). Concentrations and buffers must be optimized for each plasmid. We used 100 ng/μl of each plasmid in PBS. Typically, the best results are achieved with plasmid concentrations ranging from 0.05 to 0.1 μg/μl in PBS, Tris-EDTA or ultrapure water. Plasmids as large as ∼15 kbp can be delivered using TNT. The plasmid without the gene of interest should be used as a sham control.
In vivo TNT application
When delivering DNA via electroporation, the applied electric voltage determines how large the migration force on the charged DNA molecules is and hence how far the delivery distance will be under constant pulse duration. Simultaneously, the voltage also determines the density of pores and average pore radius on the cell membrane during electroporation. A moderate voltage (100–150 V/mm) balances the delivery distance and the pore parameters. Excessive electric voltage can damage the cell and tissue structures72,77. This protocol describes how to apply TNT on murine skin using electroporation (Steps 9–23) as well as characterization methods of plasmid delivery and gene expression (Step 25A(i–vi)). For in vivo delivery in this protocol, we used male mice aged between 8 and 12 weeks; however, we anticipate that our protocol can be applied to any strain of mice and should not be limited to a particular sex or age. For in vivo delivery experiments, it is important to deliver an empty vector control.
For all animal procedures, the animal should be anesthetized consistent with the policies and procedures of the relevant institutional laboratory animal review board. For TNT of the skin, the skin should be depilated and exfoliated to allow closer interfacing of the TNT chip with the deeper cell layers of the skin. An electric pulse is passed through a reservoir of plasmid solution and the skin, separated by a needle array–based chip. This electric pulse is a square wave with adjustable parameters. When using an electric pulse generator (CUY21EDIT II, Bex), we recommend using an initial poration pulse followed by several driving pulses. The poration pulse (200 V) can have an increased voltage to increase cell membrane pore formation, which is then followed by driving pulses (200 V) to push plasmids into the tissue. Higher voltages (>250 V) increase electrophoretic drive and create larger pores in the cell membrane, which can aid in plasmid delivery but can also be cytotoxic if excessive21,22. Pulse length is the duration the voltage is applied for that pulse, and pulse interval is the time between pulses. After TNT is administered, further characterization is required to confirm plasmid delivery and expression (Step 25).
Materials
Animals
Mice: for the results shown here, we used male C57BL/6 mice 8–12 weeks of age !CAUTION Any experiments involving animals should be undertaken only after obtaining institutional regulatory board permission and must conform to national and institutional regulations. All experimental procedures in this protocol were approved by the Institutional Animal Care and Use Committee of Indiana University (IU protocol #18047).
Reagents
Nanofabrication
4-inch double-side polished silicon wafers (doping: P-type; orientation: <100>; resistivity: 1–30 Ω cm; thickness: 525 μm; Rogue Valley Microdevices)
AZ 1518 (Integrated Micro Materials (IMM), cat. no. AZ1518MIF-G) !CAUTION AZ 1518 photoresist is an amber-red liquid with characteristic odor. It causes skin and eye irritation. High vapor concentration causes irritation to the nose, throat and lungs. Wear proper eye protection and gloves while handling and use an appropriate waste disposal container for disposal.
AZ developer 1:1 (IMM, cat. no. AZDEVEL1:1) !CAUTION AZ developer 1:1 is a clear and colorless liquid. It causes mild skin irritation and eye irritation on contact. Wear proper eye protection and gloves while handling and use an appropriate waste disposal container for disposal.
SU-8 3050 (Microchem Corp., cat. no. Y311075 O500L1GL) !CAUTION SU-8 3050 is a viscous liquid with light-yellow color. It causes skin irritation and serious eye irritation. Wear protective gloves, protective clothing, eye protection and face protection while handling and use an appropriate waste disposal container for disposal.
SU-8 developer (Microchem Corp., cat. no. Y020100 16L4PECS) !CAUTION SU-8 developer is a clear, colorless liquid. It causes mild skin irritation and serious eye irritation. Wear proper personal eye protection, clothing and gloves while handling and use an appropriate waste disposal container for disposal.
SPR 220-7.0 photoresist (Megaposit, cat. no. A016J2Q001) !CAUTION SPR 220-7.0 photoresist is a liquid with red-amber color. It causes irritation to eyes, nose and respiratory tract. Wear proper personal eye protection, clothing and gloves while handling and use an appropriate waste disposal container for disposal.
AZ 300 MIF developer (IMM, cat. no. AZ300MIF) !CAUTION AZ 300 MIF developer can cause moderate skin irritation and eye irritation on contact. Wear proper eye protection and gloves while handling and use an appropriate waste disposal container for disposal.
AZ NMP (N-methyl-2-pyrrolidone) rinse (IMM, cat. no. AZNMP) !CAUTION AZ NMP rinse is used for fast removal of photoresist. It can cause skin irritation, serious eye irritation, respiratory irritation and fertility damage after contact. Wear proper eye protection and gloves, handle in a laminar flow cabinet and use an appropriate waste disposal container for disposal.
Acetone (VWR BDH Chemicals, cat. no. BDH2025-1GLP) !CAUTION Acetone is highly volatile and flammable. It can cause skin, eye and respiratory irritation on contact. Wear proper eye protection and gloves, handle in a laminar flow cabinet and use an appropriate waste disposal container for disposal.
99.5% isopropyl alcohol (IPA; VWR BDH Chemicals, cat. no. BDH2027-1GLP) ! CAUTION IPA vapor is irritating to eyes, skin and the respiratory system. Wear proper eye protection and gloves, handle in a laminar flow cabinet and use an appropriate waste disposal container for disposal.
Hexamethyldisilazane (HMDS ; IMM, cat. no. MicroSi HMDSQ) ! CAUTION HMDS is highly flammable, toxic on skin contact and harmful if inhaled. Wear proper eye protection and gloves and handle in a laminar flow cabinet. Use an appropriate waste disposal container for disposal.
CHF3 (Matheson Tri-Gas, cat. no. HG G1643004D) for reactive-ion etcher
C4F8 (Matheson Tri-Gas, cat. no. HG G2680777D) for deep silicon etcher
SF6 (Matheson Tri-Gas, cat. no. HG G2485104D) for deep silicon etcher
Argon (Matheson Tri-Gas, cat. no. HG G1612326D) for deep silicon etcher
O2 (Matheson Tri-Gas, cat. no. HG G1678004D) for PECVD and plasma asher
N2 (Matheson Tri-Gas, cat. no. HG G1959112D) for nitrogen gun
He (Matheson Tri-Gas, cat. no. HG G1648104D) for etchers
5% (vol/vol) SiH4 (Matheson Tri-Gas, cat. no. HX G2677087DP) for PECVD
N2O (Matheson Tri-Gas, cat. no. HG G1675504D) for PECVD
Nanostrip (VWR, cat. no. 10135–756) ! CAUTION Nanostrip is irritating to eyes and skin and can cause damage to the respiratory system and central nervous system. Wear proper eye protection and gloves while handling. Handle in a laminar flow cabinet and use an appropriate waste disposal container for disposal.
Hydrofluoric acid (Fisher Scientific, cat. no. A513500) ! CAUTION Hydrofluoric acid is highly irritating to eyes, skin and the respiratory system and highly corrosive to metals. Wear proper eye protection, gloves and a face shield. Handle in a laminar flow cabinet and use an appropriate waste disposal container for disposal.
Ammonium hydroxide (Fisher Scientific, cat. no. A669C-212) ! CAUTION Ammonium hydroxide is highly irritating to eyes and skin on contact and harmful if inhaled. Wear proper eye protection and gloves. Handle in a laminar flow cabinet and use an appropriate waste disposal container for disposal.
Hydrogen peroxide (Fisher Scientific, cat. no. H3254) ! CAUTION Hydrogen peroxide is highly irritating to eyes, skin and the respiratory system and highly corrosive to metals. It can be fatal in cases of severe exposure. Wear proper eye protection, gloves and face shield. Handle in a laminar flow cabinet and use an appropriate waste disposal container for disposal.
Deionized (DI) water (Mar Cor Purification, custom)
White nitrile gloves (VWR, cat. no. 14215–762)
Polyimide film tape (McMaster Carr, cat. no. 7648A46)
UV mounting tape (Ultron Systems, cat. no. 1020R-10.0)
Large bulb pipette (8 μm) (VWR, cat. no. 16001–190)
Cleanroom swabs (Fisher Scientific, cat. no. 191600010)
Cleanroom wipes (VWR International, cat. no. 89030–168)
Tweezers (Ideal-Tek, cat. no. 1-259cf.SA)
Attaching silicon chip to reservoir
PDMS and hardener (Corning, cat. no. Sylgard 184)
Transwell 12-mm-diameter inserts (Corning, cat. no. 3460)
Petri dish (Fisher Scientific, cat. no. FB0875713A)
TNT
Isoflurane (Patterson Veterinary, cat. no. 14043070405)
Sterile Dulbecco’s PBS (Gibco, cat. no. 14190–144)
Plasmids of interest. We have successfully used fluorescent FAM-labeled DNA (IDT), Ascl1, Brn2, and Myt1l encoding plasmids (backbone, pCAGEN from Addgene plasmid #11160) constructed by Applied Biological Materials to generate the results shown here.
DI water
Ethanol wipes (Fisher Scientific, cat. no. 19-547-146)
22-26G (gauge) needle (BD)
Depilatory cream (Amazon, cat. no. B001G7PTWU)
TNT application on cutaneous tissue
Exfoliating cream (Amazon, cat. no. B002HS42UC)
3M Scotch tape (VWR, cat. no. 76247–202)
Quantifying gene expression
SuperScript III first-strand synthesis system (Thermo Fisher, cat. no. 18080051)
SYBR green PCR master mix (Fisher Scientific, cat. no. 43-091-55)
96-well reaction plate (Thermo Fisher, cat. no. 4346906)
Optical adhesive covers (Thermo Fisher, cat. no. 4360954)
0.2-ml individual tubes (Fisher Scientific, cat. no. AB-0620)
mirVana miRNA isolation kit, with phenol (Thermo Fisher, cat. no. AM1560)
Immunohistochemistry
PBS (Fisher Scientific, cat. no. BP39920)
Acetone (Sigma, cat. no. 534064) ! CAUTION Acetone is highly volatile and flammable. It can cause skin, eye and respiratory irritation on contact. Wear proper eye protection and gloves. Handle in a laminar flow cabinet and use an appropriate waste disposal container for disposal.
HardSet mounting medium (Vector Labs, cat. no. H-1400)
ImmEdge hydrophobic barrier PAP pen (Vector Labs, cat. no. H-4000)
Normal goat serum (NGS) blocking solution (Vector Labs, cat. no. S-1000)
DAPI, dihydrochloride (Thermo Scientific, cat. no. 62248)
Cryomold disposable base (Fisher Scientific, cat. no. 22-363-556)
Tissue-Tek optimal cutting temperature (OCT) compound (Electron Microscopy Sciences, cat. no. 62550–12)
Cover glasses superfrost (Fisher Scientific, cat. no. 12-542B)
Superfrost plus microscope slides (Fisher Scientific, cat. no. 1255015)
Extremus microtome blades (C.L. Sturkey, cat. no. 22210033)
Recombinant anti-beta III tubulin (Tuj1) antibody (Abcam, cat. no. ab52623; RRID: AB_869991)
Neuronal Class III beta-tubulin (TUJ1) monoclonal antibody (Covance, cat. no. MMS-435P; RRID: AB_2313773)
Goat anti-rabbit IgG H&L, Alexa Fluor 568 (Abcam, cat. no. ab175471; RRID: AB_2576207)
Goat anti-mouse IgG (H+L) cross-adsorbed secondary antibody, Alexa Fluor 568 (Thermo Fisher Scientific, cat. no. A-11004; RRID: AB_2534072)
Equipment
Cleanroom equipment
Wet bench (WAFAB International, custom)
Solvent bench (WAFAB International, custom)
Spinner (Headway Research, model no. PWM32)
Spin-rinse dryer (Laurell Tech Corp, model no. WS-650MZ-23NPPB/IND)
Hot plate (Wenesco, model no. HP1419U)
Untrasonics cleaner (Blackstone-Ney Ultrasonics, model no. multiSONIK 3)
HMDS vapor prime system (Yield Engineering Systems, model no. YES 58-TA)
Laser pattern generator (Heidelberg Instruments, model no. MLA150)
Reactive-ion etcher (Plasma-Therm, model no. Apex SLR)
Deep silicon etcher (Plasma-Therm, model no. Versaline)
PECVD (Plasma-Therm, model no. Vision 310)
Plasma asher (Richardson Electronics, Ltd., model no. Ion Wave 10)
Plasma cleaning system (Yield Engineering Systems, model no. VLF-1000)
Nitrogen gun (International Polymer Solutions, model no. Nitro-1)
Optical microscope (Nikon, model no. Eclipse L200N)
Reflectometer (Filmetrics, model no. F20 Series)
Surface Profilometer (KLA-Tencor, model no. Tencor P-7)
Scanning electron microscope (SEM; FEI, model no. Quanta 650 FEG)
Focused ion beam (Thermo Scientific, model no. Helios G4 UX Dual Beam)
Dicing saw (Disco, model no. DAD3240)
UV curing tool (Ultron Systems, model no. UH 104)
Attaching silicon chip to reservoir
Vacuum oven (Fisher Scientific, model no. 281A)
Oxygen plasma oven (Plasma Etch, model no. PE-50)
TNT
SumnoSuite low-flow anesthesia system (Kent Scientific, model no. SS-01)
Pulse generator (Bex, model no. CUY21EDIT II)
Two hook clip lead cables (Amazon, cat. no. B07TF8K9NC)
Gold-plated electrode (100 μl Neon Tip, part of kit MPK10025)
Pipettes (Thermo Fisher Scientific, cat. nos. 4642090, 4642070, 4642030 and 4642130)
Hair trimmer (Wahl, cat. no. CLP-41590 P)
Quantifying gene expression
TissueLyser (Qiagen, cat. no. 85210)
QuantStudio 3 real-time PCR instrument (Thermo Fisher)
SimpliAmp thermal cycler (Thermo Fisher, model no. A24811)
Tabletop centrifuge (Thermo Fisher, model no. 75002451)
Immunohistochemistry
Cryostat with razor (ThermoScientific, model no. CryoStar NX50)
Slide staining jar (Electron Microscopy Sciences, model no. 71385)
Slide staining rack (Electron Microscopy Sciences, model no. 71386-DR)
Confocal microscope (Olympus, model no. FV 1000)
Software
KLayout view and editor (https://www.klayout.de/)
Procedure
Fabrication of hollow-needle arrays
▲ CRITICAL The fabrication process of all three types of needles requires a cleanroom facility (International Organization for Standardization 14644–1, class 5). General guidance detailing how to do optical lithography, measure the etching rate, measure film thickness, increase etching rate, etch deeper and deposit uniform film is provided in the Supplementary Manual.
-
1Follow option A to fabricate hollow-needle arrays with flat tips (Type I), option B to fabricate hollow-needle arrays with sharp tips and center bore (Type II) and option C to fabricate hollow-needle arrays with sharp tips and off-center bore (Type III).
- Fabrication of hollow-needle arrays with flat tips (Type I) ● Timing 5 d
- Design lithography patterns with the KLayout software and save as .gds files. Three layers are required for Type I needle fabrication. An example is shown in Fig. 3a,c,d. The GDS file (TNT_type_I.gds) we use to produce the specific Type I array is available at https://doi.org/10.6084/m9.figshare.16528311.
-
Place a 4-inch Si wafer in a wafer carrier and submerge in a 50 °C Nanostrip tank for10 min to remove residual organics and metal contaminations. Follow with a DI water rinse in a quick-dump rinse tank and dry with a spin-rinse dryer for 1 min.▲CRITICAL STEP All the cleaning processes should be done on a wet bench.
- Place the wafer in a wafer carrier and put it in an HMDS vapor prime system at 110 °C for 5 min, to improve photoresist adhesion. Allow the wafer to cool down for 5 min before proceeding to the next step.
- Spin-coat one side of the wafer, defined as the backside, with an AZ 1518 photoresist at 2,000 rpm (1,000 rpm/s ramp rate) for 45 s using a spinner. Then, soft-bake the wafer on a hot plate at 110 °C for 70 s. Allow the wafer to cool down for 10 min before proceeding to the next step. The target thickness and tolerance are 2.8 μm and ±0.2 μm, respectively.
- Place the wafer in the maskless aligner (Heidelberg MLA 150) and expose at a 375-nmwavelength at a dose of 400 mJ/cm2 to define the alignment marks on the backside as shown in Fig. 3a. Two cross-alignment marks, each having a width of 20 μm and a length of 2,000 μm, are symmetrically located on a 4-inch Si wafer, both 35 mm away from the center point (Fig. 3a).
- Submerge the wafer in an AZ developer 1:1 solution in a glass beaker for 2 min with gentle agitation, followed by a DI rinse for 30 s, and spin dry for 1 min. The width of obtained alignment marks is expected to be 20 ± 1 (mean ± s.d) μm.
-
Load the wafer in a deep silicon etcher (Plasma-Therm, Versaline) to transfer the alignment marks to the Si wafer using a three-step Bosch process. First, passivate the surface with C4F8 plasma coating for 2 s using the following parameters:
C4F8 150 sccm Ar 30 sccm Chamber pressure 25 mTorr Inductively coupled plasma (ICP) power 1,000 W Bias 10 V ICP, inductively coupled plasma; standard cubic centimeters per minute (sccm), standard cubic centimeters per second.Then, perform the Etch A step for 1.5 s to remove the polymer layer from the top surface using the following parameters:SF6 100 sccm Ar 30 sccm Chamber pressure 25 mTorr ICP power 1,200 W Bias 300 V Finally, carry out the Etch B step for 2 s to etch Si using the following parameters:SF6 100 sccm Ar 30 sccm Chamber pressure 25 mTorr ICP power 1,500 W Bias 10 V Repeat this Bosch process (passivation, Etch A and Etch B) for 15 cycles, which etches ∼4 μm of silicon to create the cross-structure alignment marks as shown in Fig. 3a. The target of the etching depth and tolerance are 4 μm and ±1 μm (mean ± s.d), respectively. The tolerance of the etching profile is 90 ± 2° (mean ± s.d). The sidewall roughness is <100 nm, from the SEM image (Fig. 1k). - Remove the residual AZ 1518 photoresist by dipping in acetone in a glass beaker, sonicate for 10 min in an ultrasonic cleaner rinse with IPA for 1 min and spin dry for 1 min. Then, dry the wafer at 150 °C for 10 min on a hot plate and allow the wafer to cool down for 5 min.
- Place the wafer in a plasma cleaning system (YES VLF-1000) and run an oxygen plasma cleaning to remove the residual organics for 60 s using the following parameters:
O2 100 sccm Chamber temperature 25 °C Chamber pressure 10 mTorr Power 100 W -
Spin-coat a negative photoresist of SU-8 3050 on the backside (the same side as the alignment marks) at 500 rpm (100 rpm/s ramp rate) for 10 s and 3,000 rpm (1,000 rpm/s ramp rate) for 30 s (Fig. 4a). Then, relax the wafer on a flat aluminum plate for 10 min to allow the SU-8 3050 to evenly distribute. Then, soft-bake at 95 °C for 12 min on a hot plate. The film thickness can be measured by a refractometer (Filmetrics, model no. F20 series). The target thickness and tolerance are 50 μm and ±2 μm, respectively.▲ CRITICAL STEP Bubbles or particles can cause inhomogeneous photoresist coating that results in non-uniform patterning. The coating quality can be easily seen by the naked eye by tilting the wafer. If the coating is inhomogeneous, remove the SU-8 3050 photoresist with SU8 developer and redo the process from Step 1A(ix). A nitrogen gun can be used to blow away particles on the wafer surface before spin coating to improve the homogeneous coating. Meanwhile, keep the wafer on a flat, level surface for 10–20 min so that SU-8 photoresist can re-flow to improve the uniformity.
-
Load the wafer in the maskless aligner. Move the stage in X and Y directions to find the two alignment marks using the top camera and measure the center positions for each cross mark from the camera images. Then, expose at a 375-nm wavelength at a dose of 450 mJ/cm2 to define the backside of the hollow channels, which has arrays of 20-μm diameter with 150-μm spacing (Fig. 3c). Post-bake the wafer at 95 °C for 5 min on a hot plate and move the wafer onto a cleanroom wipe on a table and allow it to cool down for 10 min.▲ CRITICAL STEP This step requires precise alignment. If the alignment is out of the tolerance, the device cannot be fabricated because of misalignment of the back- and front-side etching. Therefore, getting a clear and sharp image for each mark is important to measure accurate positions. The images can be adjusted by the camera settings, such as focus, brightness, contrast and zoom. The tolerance is ±1.0 μm.
-
Place the wafer on a hot plate, raise the temperature from room temperature (24 °C) to 250 °C and maintain at 250 °C for 60 min to hard-bake (ramp rate = 450 °C/h; cool-down rate = 450 °C/h). The SU-8 photoresist should turn brown. See Fig. 5b.▲CRITICAL STEP Although this step enhances the etching selectivity over Si, it also prevents the SU-8 photoresist from sticking to the ceramic clamp in the chamber (Plasma-Therm, model no. Versaline) during the deep silicon-etching process.
-
Place the wafer (Fig. 5c) in a plasma asher and run an oxygen plasma cleaning (O2 = 250 sccm; power = 600 W; cleaning time: ≥1 h) until the brown color has disappeared.▲ CRITICAL STEP The oxygen plasma is a more effective way to remove the hard-baked SU-8 compared to cleaning in Nanostrips or Piranha solution, which takes more time.
- Dip the wafer in a 50 °C Nanostrip tank for 30 min to remove residual organics and metal contamination. Rinse the wafer with DI water for 5 min, submerge it in 2% (wt/vol) hydrofluoric acid in a PTFE (polytetrafluoroethylene; Teflon) beaker for 2 min, rinse the wafer with DI wafer for 5 min and spin dry for 1 min. See Fig. 5d.
- Place the wafer with the backside facing up on a hot plate at 150 °C for 10 min to dry andcarry out the HMDS prime, as described in Step 1A(iii).
-
Spin-coat the front side with a positive photoresist (SPR 220-7.0) at 3,000 rpm (1,000 rpm/s ramp rate) for 30 s (Fig. 4d). Then, pre-bake the wafer at 115 °C for 90 s on a hot plate. The target thickness is 7.5 μm, with a tolerance of ±0.3 μm.▲ CRITICAL STEP Move the wafer onto cleanroom wipes on a table to let it gradually cool down for 10 min, to prevent the photoresist from cracking.
-
Load the wafer in the maskless aligner. Move the stage to find the alignment marks on the backside using the bottom camera and measure the center positions and expose at a 375-nm wavelength with a dose of 400 mJ/cm2 to define the pattern (Fig. 3d) on the front side, which has arrays of donut-like shapes (50–70-μm outer diameter and 5–15-μm inner diameter) with 150-μm spacing. The tolerances of the generated pattern are ±3 μm for the outer diameter and ±1 μm for the inner diameter.▲CRITICAL STEP This step requires precise alignment using the cross marks on the backside. If the marks are contaminated or blurred (out of focus because of large particles on the backside), use cleanroom wipes to wipe down the area with acetone and IPA and blow-dry it immediately with the nitrogen gun. Clear and sharp images are required to minimize alignment errors. The tolerance of the alignment is ±1.0 μm.
-
Post-bake the wafer at 115 °C for 90 s on the hot plate. Move the wafer from the hot plate directly onto cleanroom wipes and leave for 30 min.▲CRITICAL STEP This post-bake process requires a waiting time of ≥30 min after exposure. Cool down the wafer slowly to prevent the photoresist cracking.
- Submerge the wafer in the AZ 300 MIF developer in a glass beaker for 3 min and 30 s, rinse the wafer with DI water for 30 s and spin dry it for 1 min. See Fig. 4e.
-
Prepare several cleanroom wipes on a table as a cushion and then gently place the wafer on the wipes with the backside facing up to prevent damage on the patterned front side. Cover the etched hollow microchannel arrays on the backside with polyimide film tapes (Fig. 4f and Fig. 5i,j). Gently roll a cleanroom swab over the polyimide tape to minimize possible air gaps.▲CRITICAL STEP The hollow channels on the backside must be taped with the polyimide film. Otherwise, helium gas that is used to cool the wafer during the etching will leak into the chamber when the wafer is etched through, which can cause equipment errors.
-
Load the wafer in the deep silicon etcher with the patterned front side facing up. Perform another Bosch process. Run the passivation step for 2 s using the following parameters:
C4F8 100 sccm Ar 30 sccm Chamber pressure 25 mTorr ICP power 1,000 W Bias 10 V Next, run Etch A for 1.5 s using the following parameters:SF6 100 sccm Ar 30 sccm Chamber pressure 25 mTorr ICP power 1,200 W Bias 300 V Finally, run Etch B for 2 s using the following parameters:SF6 100 sccm Ar 30 sccm Chamber pressure 25 mTorr ICP power 1,500 W Bias 10 V Repeat the above Bosch process (passivation, Etch A and Etch B) for 1,500 cycles, to create ∼210-μm-long needle arrays. The needle heights can be measured using a Surface Profilometer (KLA-Tencor, P-7). The target etching depth is 210 μm, and the tolerance is ±10 μm. At the end of the process, the wafer should be etched through, and hence light will pass through from the backside to the front side (Fig. 2e and Fig. 4f).▲CRITICAL STEP Frequently stop (∼every 20 cycles) the Bosch etch process after 1,400 cycles and monitor the wafer under an optical transmission microscope (Nikon, Eclipse L200N) to check if the hollow channels are etched through. If the channels are etched through, light will pass through from the backside to the front side (Fig. 2, b and e). Otherwise, additional etching will be required until holes are visible under the optical transmission microscope (Fig. 2, c and f). The tolerance for this process is >95% of the holes etched through. - Gently remove the backside polyimide film tapes using plastic tweezers to avoid scratches.
- Submerge the wafer in the AZ NMP rinse solution in a glass beaker at 80 °C for 2 h to remove the residual SPR 220-7.0 photoresist. Then, rinse the wafer in DI water for 5 min and spin dry for 1 min.
- Place the wafer in a wafer carrier and emerge it in a 50 °C Nanostrip tank for 30 min to remove the residual organics and metal contaminations, rinse with DI water, dip in hydrofluoric acid for 2 min, rinse with DI water for 5 min and spin-dry it for 1 min. See Fig. 4g.
-
Place the backside of the wafer on a UV mounting tape. Dice the wafer into 17-mm-by-17-mm squares (or alternative appropriate sizes) with a dicing saw system (Disco, model no. DAD3240). After dicing, illuminate UV light on the backside for 1 min in a UV curing system (Ultron Systems, model no. UH 104), and then gently take out the diced TNT chips from the UV tape. See Fig. 5, k and l.▲CRITICAL STEP Prepare several cleanroom wipes on a table and place the front side (needle side) face down onto the cleanroom wipes. Gently place a UV tape with a metal frame to the backside and gently roll over the wafer with a cleanroom swab to remove air gaps, so that the wafer is well mounted onto the UV tape.
- Clean the diced TNT chips in acetone in a glass beaker, sonicate with weak power (∼5 W) for 1 min in the ultrasonic cleaner and rinse with IPA for 1 min and DI water for 1 min. Then, dip in Nanostrip in a glass beaker at 50 °C for 10 min to remove residual organics and metal contaminations and rinse with DI water for 5 min. Finally, clean the chips in a glass beaker with the standard RCA SC1 (ammonium hydroxide/hydrogen peroxide/H2O = 1:1:5 (vol/vol) at 80 °C for 10 min) to remove particles, rinse with DI water for 5 min and blow dry with the nitrogen gun. See Fig. 5l. The needle side should always be face up during cleaning procedures, and handling should be gentle to prevent damage.
-
Measure the bore sizes with an SEM (FEI, model no. Quanta 650 FEG). Then, place the TNT chips in a PECVD system and deposit a SiO2 film to shrink the bore size to the target size using the following parameters:
Chamber temperature 300 °C SiH4 100 sccm O2 50 sccm Power 300 W Deposition rate 75 nm/min ▲CRITICAL STEP Because the deposition rate and the bore size shrink rate are different, the actual shrink rate must be measured using SEM imaging (Supplementary Manual). The final bore size is an important factor that affects TNT performance, because it affects the amount of DNA delivered to the tissue. If the bore size is <3 μm, the oxide layer can be removed by hydrofluoric acid solution, and Step 1A(xxix) can be repeated.
- Fabrication of hollow-needle arrays with sharp tips (Type II) ● Timing 6 d
- Design lithography patterns with the KLayout software and save as .gds files. Four kinds of layers are required for Type II needle fabrication as shown in the example in Fig. 3a, c–e. The GDS file (TNT_type_II.gds) we use to produce the specific Type II array is available at https://doi.org/10.6084/m9.figshare.16528311.
- Follow Step 1A(ii–xvi) (the fabrication process for the Type I needle).
-
Place the wafer with front side facing up in the PECVD system and deposit a 350-nm-thick SiO2 film using the following parameters:
Chamber temperature 300 °C SiH4 100 sccm O2 50 sccm Power 300 W Deposition rate 75 nm/min The target thickness of the oxide is 350 nm, and the tolerance is ±15 nm. - Perform the HMDS priming as described in Step 1A(xvii).
- Spin-coat AZ 1518 photoresist on the front side at 2,000 rpm (1,000 rpm/s ramp rate) for45 s and soft-bake at 110 °C for 70 s on a hot plate. The AZ 1518 is used as an etching mask to etch the circle arrays. The target thickness is 2.8 μm, with a tolerance of ±0.2 μm. See Fig. 7a.
-
Load the wafer in the maskless aligner. Move the stage to find the cross-alignment marks on the backside using the bottom camera to measure the center positions and expose at a 375-nm wavelength at a dose of 400 mJ/cm2 to define the disk arrays (disk diameter = 50–70 μm; spacing = 150 μm). See Fig. 3e.▲CRITICAL STEP This step requires precise alignment using the cross marks on the backside. If the marks are contaminated or blurred because the wafer is tilted due to particles on the backside, use cleanroom wipes to wipe down the area with acetone and IPA and blow-dry it with the nitrogen gun. Clean and clear images of the marks are essential to minimize the alignment error. The tolerance of the alignment is ±1.0 μm.
- Submerge the wafer with the front side facing up in the AZ developer 1:1 solution in a glass beaker for 2 min with gentle agitation, followed by a DI rinse for 30 s, and spin dry for 1 min. The target diameter of the circle is 55 μm, and the tolerance is ±3 μm. See Fig. 7b.
-
Place the wafer in a reactive-ion etcher (Plasma-Therm, model no. Apex SLR) to etch the deposited oxide layer using the following parameters:
CHF3 50 sccm Chamber pressure 20 mTorr ICP 500 W Bias 50 W SiO2 etch rate 70 m/min See Fig. 7c. Measure the oxide thickness with the Filmetrics and make sure that it is completely etched.▲CRITICAL STEP This step etches away the 350-nm-thick oxide layer on the surface except the disk arrays covered by the patterned AZ 1518. - Place the wafer in acetone in a glass beaker, sonicate for 10 min to remove the residual AZ1518, rinse in IPA for 1 min and spin-dry it for 1 min. Repeat this step to completely remove the photoresist, if needed.
- Place the wafer on a hot plate at 150 °C for 10 min, with the front side facing up.
- Prime with HMDS as described in Step 1A(iii).
- Spin-coat the front side with SPR 220-7.0 photoresist at 3,000 rpm (1,000 rpm/s ramp rate) for 30 s. See Fig. 7d.
-
Pre-bake the wafer at 115 °C for 90 s on a hot plate. Move the wafer from the hot plate on to cleanroom wipes and let it cool down for 10 min.▲ CRITICAL STEP Cooling is required to prevent the photoresist from cracking. Photoresist cracking is easily seen by the naked eye. If it cracks, dip the wafer in AZ 300 MIF developer for 5 min to completely remove the photoresist and redo the step from Step 1A(xvii).
-
Load the wafer in the maskless aligner. Move the stage to find the cross-alignment marks on the backside using the bottom camera to measure the center positions and expose at a 375-nm wavelength with a dose of 400 mJ/cm2 to define the pattern of hollow needles, which has arrays of donut-like shapes (50–70-μm outer diameter and 5–10-μm inner diameter) with 150-μm spacing. See Fig. 3d and Fig. 7e. The tolerance of the outer diameter and inner diameter are ±3 μm and ±1 μm, respectively.▲ CRITICAL STEP This step requires precise alignment using the cross marks on the backside. If the marks are contaminated or blurred because the wafer is tilted due to particles on the backside, use cleanroom wipes to wipe down the area with acetone and IPA and blow-dry it with the nitrogen gun. Clean and clear images of the marks are essential to minimize alignment error. The tolerance of the alignment is ±1.0 μm.
-
Post-bake the wafer at 115 °C for 90 s on a hot plate. Leave the wafer for ≥30 min after exposure to cool down.▲ CRITICAL STEP The wafer must be cooled slowly to prevent the photoresist from cracking.
- Submerge the wafer in the AZ 300 MIF developer in a glass beaker for 3 min and 30 s, rinse the wafer with DI water for 30 s and spin-dry it for 1 min. See Fig. 7e.
-
Prepare several cleanroom wipes on a table as a cushion and then gently place the wafer on the wipes with the backside facing up to prevent damage on the patterned front side. Then, tape the etched channel arrays on the backside with polyimide film tapes (Fig. 7f and Fig. 5i). Gently roll a cleanroom swab over the polyimide tape to minimize possible air gaps.▲ CRITICAL STEP The hollow channels on the backside must be taped with the polyimide film. Otherwise, helium gas that is used to cool the wafer during the etching will leak into the chamber when the wafer is etched through, which can cause equipment errors.
-
Place the wafer in the deep silicon etcher with the front side facing up and perform isotropic etching to form sharp tips. Run the passivation step for 1 s using the following parameters:
C4F8 100 sccm Ar 30 sccm Chamber pressure 25 mTorr ICP power 1,000 W Bias 10 V Next, run Etch A for 1 s using the following parameters:SF6 500 sccm Ar 30 sccm Chamber pressure 25 mTorr ICP power 1,200 W Bias 300 V Finally, run Etch B for 30 s using the following parameters:SF6 500 sccm O2 30 sccm Chamber pressure 25 mTorr ICP power 2,500 W Bias 10 V Repeat the process (passivation, Etch A and Etch B) for 30 cycles to create the sharper tip. See Fig. 7f and Fig. 8c,f.▲ CRITICAL STEP The isotropic Si etching forms the sharp tips, and the tip size reduces over etching time. Frequently monitor the tip size by taking out the wafer from the chamber and observing it under an optical microscope to ensure that the tip is not over-etched. The tip size should gradually decrease over etching time as shown in Fig. 8. Move to the next step when the tip size is reduced to the target value. Use the optical microscope to measure the tip sizes. The target tip size is ∼10 μm, with a tolerance of ±2 μm. -
Place the wafer in the reactive-ion etcher (Plasma-Therm, model no. Apex SLR) toetch the oxide layer remaining in the holes at the center of the tips using the following parameters (Fig. 7g):
CHF3 50 sccm Chamber pressure 20 mTorr ICP 500 W Bias 50 W SiO2 etch rate 70 nm/min ▲ CRITICAL STEP The 350-nm-thick oxide layer at the center of the disk must be etched away so that the etching gas can penetrate the oxide hole to etch under the Si substrate to form hollow channels.
- Fabrication of hollow-needle arrays with sharp tips and off-center bore (Type III) ● Timing 6 d
- Design lithography patterns with the KLayout software and save as .gds files. Four layers are prepared for Type III needle fabrication as shown in the examples included in Fig. 3, a, c, e and f. The GDS file (TNT_type_III.gds) we use to produce the specific Type III array is available at https://doi.org/10.6084/m9.figshare.16528311.
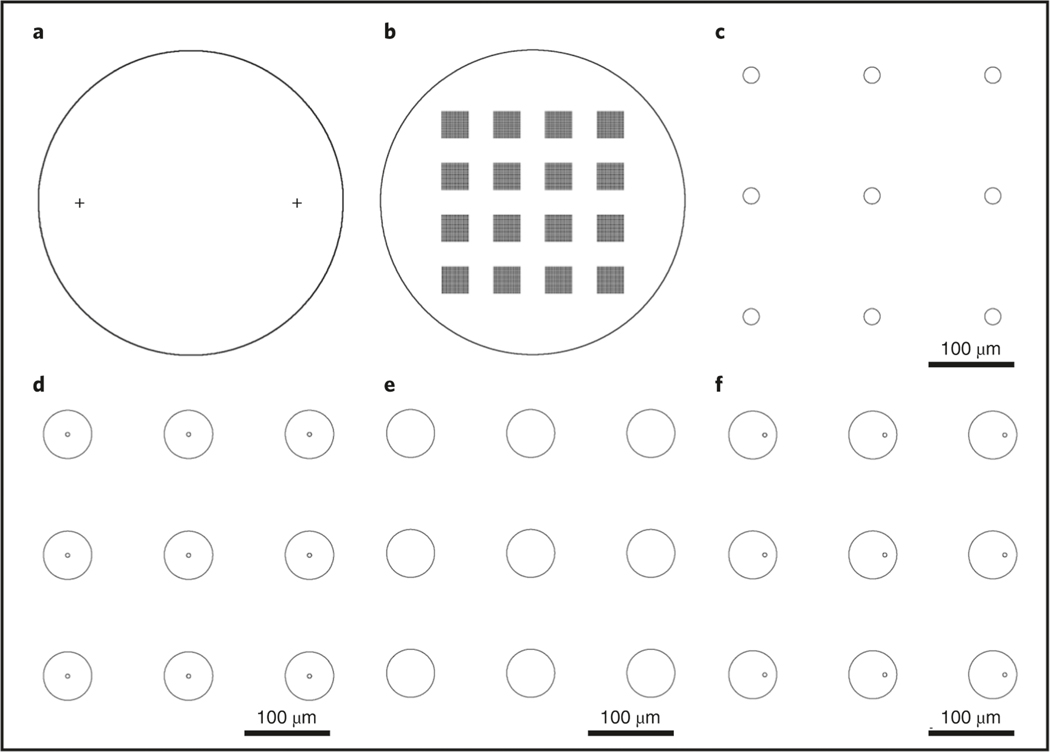
Fig. 3 |. Typical lithography patterns for needle arrays in the fabrication process.
a, Two alignment marks, each having a width of 20 μm, are symmetrically located on a 4-inch Si wafer, both 35 mm away from the center point. b, 4 by 4 chips are arranged on a 4-inch Si wafer with a 17-mm period. Each chip has a 60 by 60 array of the needles with a 150-μm period (distance between the needles on the chip). c, The backside etching mask of holes with the size of 20 μm, with a 150-μm period. d, The layout of the primary Type I needle array with a 55-μm diameter and a 150-μm period; the center bore size is 5 μm. e, The layout array with a 55-μm diameter and a 150-μm period used to generate the SiO2 protection layer in the isotropic etching step of the Type II needle. The layout in d is reused in the fabrication process of the Type II needle array. f, The layout of the Type III needle array with 55-μm diameter and 150-μm period; the 5-μm bore is positioned 15 μm from the center. The layout in e is reused in the fabrication process of the Type III needle array.
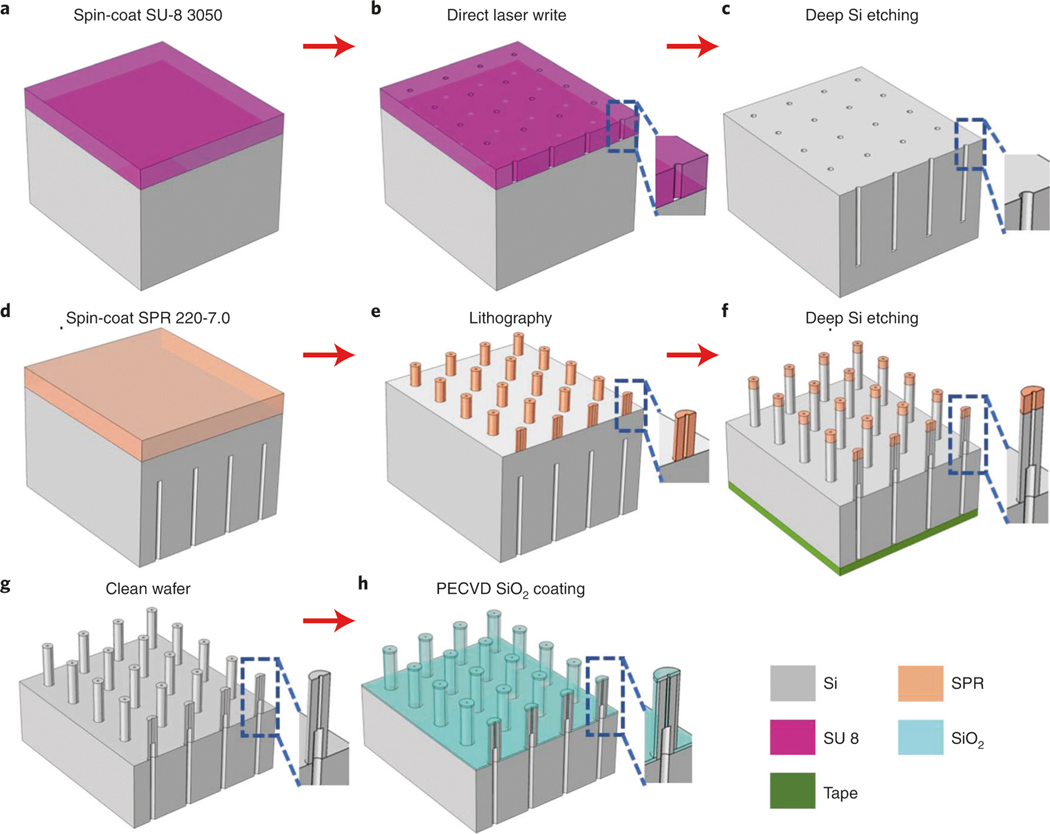
Fig. 4 |. Schematics of the fabrication process of the primary hollow needles with flat tip (type I).
a, Spin-coat an ∼50-μm-thick SU-8 3050 photoresist on the backside of the silicon wafer (Step 1A(x)). b, Pattern the layout (Fig. 3c) using the maskless aligner, followed by hard baking at 250 °C for 1 h (Step 1A(xi–xiii)). c, Etch deep channels on the wafer using the deep Si etcher (Step 1A(xiv)). d, Remove the residual SU-8 photoresist and spin-coat SPR 220-7.0 photoresist on the front side (Step 1A(xv–xviii)). e, Pattern the donut-shaped arrays (Fig. 3d) (Step 1A(xix–xxi)). f, Cover the backside holes using polyimide film and etch the hollow-needle arrays (Step 1A(xxii–xxiii)). g, Clean and dice the wafer (Step 1A(xxiv–xxviii)). h, Shrink the bore size to the target value using PECVD SiO2 deposition (Step 1A(xxix)).
Fig. 5 |. Etching and dicing processes.
a, Hole arrays were patterned in SU-8 3050 photoresist using the maskless aligner. b, Hard-bake at 250 °C for 1 h. c, Deep Si etch with the Bosch process. d, Remove the residual SU-8 photoresist. e, Optical images of SU-8 3050 photoresist with 20-μm hole arrays. f, the SU-8 photoresist turned a brown color after the hard bake at 250 °C. g and h, Residual SU-8 photoresist after the Bosch process of 430-μm-deep Si etching (g) and after cleaning the residual SU-8 photoresist (h). Images were taken from the top view. i, Polyimide film tape is used to cover the holes on the backside before etching the front side. j, A zoomed-in optical image of i showing that the holes are covered with the tape. k, 4 × 4 TNT chips on a 4-inch Si wafer with 17-mm period. l, Diced TNT chips. Scale bars, 300 μm.
Fig. 6 |. Controlling hole size with oxide deposition.
a and b, SEM images of silicon hollow needles before (a) and after (b) the deposition of SiO2 by PECVD. Zoomed-in views are given in c and d, in accordance with selected areas in a and b, respectively. The bore size is shrunk down from 5.8 μm (c) to 2.5 μm (d). Images were taken from the top view. Scale bars, a and b, 100 μm; c and d, 5 μm.
Fig. 7 |. Schematics of the fabrication process for the hollow needles with sharp tip and centered bore (Type II).
a, Spin-coat AZ 1518 photoresist on the front side with PECVD SiO2 coating (Step 1B(iii–v)). b, Expose the layout (Fig. 3e) using the maskless aligner followed by development (Step 1B (vi–vii)). c, Etch away the SiO2 layer without the photoresist covering using DRIE (Step 1B(viii)). D, Remove the residual AZ 1518 and spin-coat SPR 220-7.0 photoresist (Step 1B(ix–xiii)). E, Pattern the donut-shaped arrays (Fig. 3d) (Step 1B(xiv–xvi)). F, Cover the backside holes using polyimide tape and carry out isotropic Si etching to obtain sharp tips (Step 1B(xvii–xviii)). g, Etch through the SiO2 layer in the center to expose the Si underneath (Step 1B(xix)). h, Form the hollow-needle array with the deep Si etcher (Step 1A(xxiii)). i, Remove the residual photoresist and oxide, followed by cleaning the wafer (Step 1A(xxiv–xxix)).
Fig. 8 |. Isotropic etching of Si to fabricate the sharp needle (Type II).
a–c, Optical images of the needles before sharpening with isotropic etching (a), after etching in cycle 1 (b) and after etching in cycle 2 (c). Selected areas in a–c are zoomed in and shown in d–f, respectively. Scale bars, a–c, 200 μm; d–f, 50 μm.
Fig. 9 |. Schematics of the fabrication process for the hollow-needle array with sharp tip and off-center bore (Type III).
a, Spin-coat AZ 1518 photoresist on the front side of the Si wafer with PECVD SiO2 deposition. b, Expose the layout (Fig. 3e) using the maskless aligner followed by development. c, Etch away the SiO2 layer in the patterned area. d, Remove the residual AZ 1518 and spin-coat SPR 220-7.0 photoresist. e, Pattern the disk arrays with an off-centered hole (Fig. 3f). f, Cover the backside holes using polyimide tape and carry out isotropic Si etching to obtain sharp tips. g, Etch through the SiO2 layer to open the off-centered holes. h, Form the off-centered hollow-needle array using the Bosch process. i, Remove the residual photoresist and oxide, followed by cleaning the wafer. See Step 1C(i–ii) for details of all the processes.
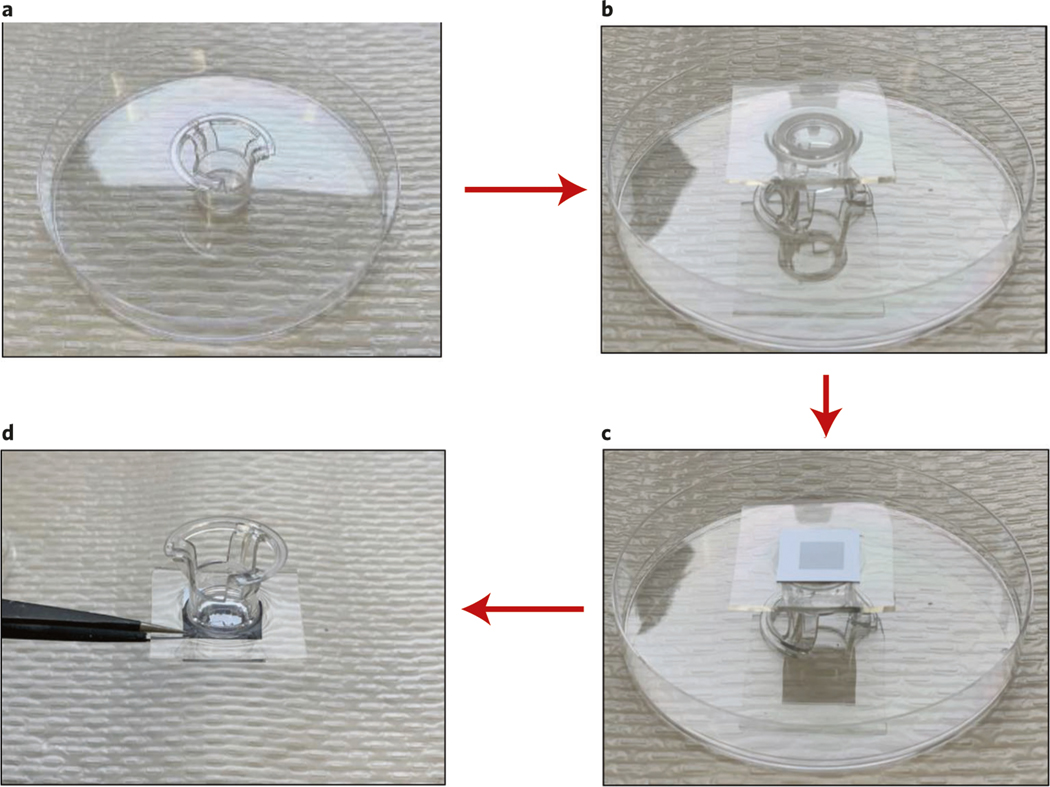
Attaching the silicon chip to the reservoir ● Timing 5 h
-
2
Mix PDMS and hardener at a 10:1 ratio (∼5 ml) and degas in a vacuum oven (67.7 kPa) for 30 min.
-
3
Cut the membrane off the bottom of a Transwell, which will act as the reservoir when attached above the nanofabricated chip.
-
4
Pour PDMS into a Petri dish, which acts as a mold. Insert the Transwell into the PDMS solution. The PDMS volume should be enough to cover the side walls. See Fig. 10a.
-
5
Cure the sample in a 60 °C oven for 4 h. See Fig. 10a.
-
6
Using a razor, cut out the PDMS at the bottom of the Transwell from inside. See Fig. 10b.
-
7
Place the cured PDMS with embedded Transwell in an oxygen plasma oven with the nanofabricated chip for 2 min to fully bond the PDMS layer and chip. See Fig. 10c.
-
8
Place the chip in an 80 °C oven for another 45 min to fully cure and stabilize the PDMS layer. Then, cut away any extraneous PDMS. See Fig. 10d.
■ PAUSE POINT For storage, secure chip needles facing up in a box without the needles contacting any walls and keep at room temperature. The chip can be stored in a mechanical shock–free environment at room temperature for several years.
Fig. 10 |. Preparation of the reservoir-mounted chip (Steps 2–8).
a, Cast and cure PDMS with embedded reservoir. b, Cut out the PDMS at the bottom of the reservoir from inside. c, Attach PDMS with embedded reservoir to the chip using plasma curing. d, Final chip with attached reservoir.
TNT (mouse skin) ● Timing 15 min
! CAUTION Any experiments involving animals should be undertaken only after obtaining institutional regulatory board permission.
▲ CRITICAL Dehairing of the site at which TNT is to be applied must be undertaken 24–48 h before applying TNT.
-
9
Anesthetize the mouse by following the procedure approved by your institution. We use continuous administration of 1–3% (vol/vol) isoflurane.
-
10
Shave the skin to be transfected with hair trimmers.
-
11
Apply depilatory cream to the shaved area for 30–60 s and then remove it using water and paper towels.
-
12
Remove the mouse from anesthesia and allow it to regain consciousness before returning it to its cage.
-
13
On the day of the TNT procedure (i.e., 24–48 h later), anesthetize the mouse by following the procedure approved by your institution. We use continuous administration of 1–3% (vol/vol) isoflurane.
-
14
To remove the top dead layers of the epidermis and aid in cargo delivery penetration, tape-strip the dehaired area six times with Scotch tape. Wipe clean and then rub exfoliating cream in a circular motion on the area for 1 min. Clean the area with water and then alcohol wipes.
▲ CRITICAL STEP Cleaning and eliminating the upper layers of the epidermis through tape stripping and exfoliating cream are important when trying to transfect cells in the dermis. Overexfoliating should be avoided because this can lead to removal of deeper layers of the skin, cause minor wounds and induce inflammation.
-
15
Slide a 26G needle intradermally where the skin will be transfected.
-
16
Using the hook clip wire, connect the 26G needle to the positive terminal of the pulse generator.
-
17
Using a 1-ml pipette, add 500 μl of cargo (plasmid) delivery solution (∼100 ng/μl in PBS) to the reservoir of the nanofabricated chip.
▲ CRITICAL STEP The appearance of liquid outside the reservoir indicates leakage between the reservoir and the PDMS seal. If leakage is observed, then that chip should not be used for TNT.
-
18
Place the nanofabricated chip directly on the skin so that the 26G needle passes through the middle of the area covered by the chip. Press the chip firmly down by hand to ensure contact throughout the procedure.
▲ CRITICAL STEP The chip must remain in contact with the skin throughout the electroporation process. This requires applying firm pressure (but not hard enough to break the chip) to avoid loss of contact due to any movement caused by the electric pulses, which vary by TNT location.
-
19
Submerge the gold-plated electrode, connected to the negative terminal of the pulse generator, in the plasmid solution in the reservoir on top of the nanofabricated chip. The tip should be just covered by the solution, and it should not touch the chip.
-
20Apply a square electric pulse. The TNT pulse generator parameters that we use are in the following table:
Parameter Unit Driving pulse (Pd) maximum current 300 mA Poration pulse voltage 200 V Poration pulse length 10 ms Poration interval 100 ms Pd voltage 200 V Pd length (Pd on) 10 ms Pd interval (Pd off) 100 ms Number of pulses (Pd cycle) 10 Decay rate 0% -
21
Remove the nanofabricated chip and 26G needle carefully.
-
22
Remove the mouse from anesthesia and allow it to regain consciousness before returning it to its cage.
-
23
Clean the chip by removing the cargo solution and then dipping it in DI water and then in 100% ethanol and store it in an airtight plastic container at room temperature (15–30 °C) until next use.
Evaluation of TNT effect on the animal ● Timing Variable depending on technique
▲ CRITICAL In our experience, an impact on gene expression can be detected by PCR and immunohistochemistry by day 1 and day 3 after TNT, respectively.
-
24
Euthanize the animal according to your institutional animal regulatory board guidelines at an appropriate time point after TNT application. Collect the skin samples from the area where TNT was performed. Snap-freeze skin samples in liquid nitrogen for gene expression analysis. For immunohistochemistry, collect skin samples either in 4% (vol/vol) paraformaldehyde for paraffin sectioning or in OCT for cryosectioning.
-
25Follow option A to quantify gene expression and/or option B to undertake immunohistochemistry to evaluate changes in protein expression after TNT.
- Quantifying gene expression ● Timing 7 h
-
Collect the transfected tissue and place it immediately in a cryovial in liquid nitrogen.■ PAUSE POINT Samples can be stored at −80 °C indefinitely and further processed later.
- Without thawing the tissue, grind it in liquid nitrogen with a mortar and pestle until it becomes a powder.
- Add lysis buffer from a MirVana total RNA isolation kit. Use a volume that is appropriate for the tissue weight according to the manufacturer’s recommendation.
- To further dissociate the tissue, use the TissueLyser. Use 30 Hz for 12 s, with three intervals with 10 s in between.
- Extract the RNA using the MirVana total RNA isolation kit and following the manufacturer’s protocol.
-
Reverse-transcribe the RNA into cDNA using Oligo(dT)20 primers. Use quantitative reverse-transcription PCR (RT-qPCR) with primers for your gene of interest to quantify the expression of your gene.
-
- Immunohistochemistry ● Timing 2 d
- Dissect tissue and place it into cryomolds with OCT.
- Immediately flash-freeze on an aluminum foil boat floating on liquid nitrogen.
- Cryosection the tissue (10-μm-thick sections).
- Wash with PBS to remove OCT and then fix in ice-cold acetone for 5 min.
- Wash sections in PBS for 3 min (three to five dips) followed by 1.5 h of blocking in 10% (vol/vol) NGS.
- Incubate tissue overnight at 4 °C in an appropriate concentration of primary antibody in2.5% (vol/vol) NGS in PBS with 0.1% (vol/vol) Tween 20. For example, we used Tuj1 (Neuronal Class III beta-tubulin monoclonal antibody) at a dilution of 1:500 for Fig. 11d.
- Wash tissue in PBS (5 min) and incubate with an appropriate concentration of fluorescent antibody for 1 h at room temperature. To detect Tuj1, we used goat anti-mouse IgG Alexa Fluor 568 antibody at 1:200 dilution for Fig. 11d.
- Wash tissue in PBS (5 min) and then incubate in an appropriate concentration of DAPI(we used a 1:10,000 dilution) for 2 min.
- Wash samples (5 min) and mount coverslips.
- Image tissue. For the images shown in Fig. 11d, we used a laser scanning confocal microscope (Olympus FV 1000 spectral).
Fig. 11 |. Cutaneous TNT and in vivo reprogramming.
a, Schematic representation of TNT of dorsal mouse skin. b, Image of a mouse undergoing TNT. c, Gene expression of skin 24 h after TNT with ABM (Ascl1, Brn2 and Myt1l) shows increased expression of delivered genes. Data expressed as mean ± s.d. *P < 0.05 (n = 4). d, 3 weeks after TNT with ABM, skin was collected and stained for neuronal marker Tuj1 (red) and DAPI (blue) and imaged using confocal microscopy. Tuj1+ neurites are seen extending from a cell body (identified by the nucleus), indicating an induced neuron. Neuron cell bodies are not naturally found in the skin.
Troubleshooting
Troubleshooting advice can be found in Table 1.
Table 1 |.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
|
| |||
| 1A(xi) | Patterned sizes are out of the tolerance | The exposure time and development time are not optimized | Spin SU-8 3050 on a dummy Si wafer and perform the dose test to find out the correct parameters |
| 1A(xiv) | The etching depth did not reach the target of 430 ± 10 μm | Residual SU8 resist remained in the holes because of insufficient development time in Step1A(xii), or the etch rate is changed because of buildup of polymers in the chamber | Use a Filmetric with a smaller beam size (≤10 μm) to measure thicknesses in the holes. If SU8 remains in the holes, etch it away with oxygen plasma. Alternatively, run an oxygen plasma cleaning and then continue etching the wafer until it reaches the target depth |
| 1A(xv) | SU8 is not etched | The plasma tool setting is not optimized to remove the hard-baked SU8 | Increasing the plasma power, chamber temperature and oxygen flow rate will increase the SU8 ashing rate |
| 1A(xix) | The alignment error is out of the tolerance | The alignment marks are contaminated, and/ or there are particles on the backside | Carefully wipe down the alignment marks using wet cleanroom papers with acetone followed by a nitrogen blow-dry until all the alignment marks can be seen clearly |
| 1A(xxiii) | Some of the hollow microchannels are blocked or not opened yet | The etching time is not long enough, or contamination and particles exist in some holes | Etch until the clogged channels are opened. Keeping the fabrication environment clean and avoiding contamination from particle sources are important |
| 1A(xxix) | The bore size is smaller than the target size after PECVD oxide deposition | Over-deposited SiO2 layer | Remove the coated oxide layer in hydrofluoric acid solution and redeposit the PECVD oxide layer (Step 1A(xxix)) to get the target bore size |
| 9 and 13 | The animal is agonal breathing | Anesthesia is too deep, or the flow rate is not strong enough to circulate enough air | Lower the isoflurane concentration or increase the air flow rate |
| 20 | No electric current (absence of convulsions) | Electrical issue, chip blocked or loss of contact between the chip and the skin | Check wires and connections, clean the chip through vigorous pipetting of water, use a new chip or maintain firm pressure on the chip against the skin |
| 25A(vi) | Low gene expression | Not enough plasmid was delivered, or the plasmid was not transfected deeply enough | Increase the plasmid concentration, or increase the number of pulses or the voltage |
Timing
Step 1A(i–xxix), fabrication of hollow-needle arrays with flat tips (Type I): 5 d
Step 1B(i–xx), fabrication of hollow-needle arrays with sharp tips and center bore (Type II): 6 d
Step 1C(i–ii), fabrication of hollow-needle arrays with sharp tips and off-center bore (Type III): 6 d
Steps 2–8, attaching the silicon chip to the reservoir: 5 h
Steps 9–23, TNT of mouse skin: 15 min
Step 24, euthanasia and collection of skin samples: variable
Step 25A(i–vi), quantifying gene expression: 7 h
Step 25B(i–x), immunohistochemistry: 2 d
Anticipated results
TNT chip fabrication
The procedures described in this protocol enable fabrication of three types of silicon hollow-needle arrays. We have successfully fabricated and used needles with outer diameters of 50–90 μm, inner diameters of 5–15 μm and backside hole sizes of 20–50 μm. Thus, the design parameters can be chosen on the basis of the capability of the tools available for fabrication.
Figure 1, h and k show the SEM images of fabricated Type I flat-tipped Si hollow needles in a uniform array and zoomed-in view, respectively. In this example, the length of a cylindrical needle is ∼210 μm, the outer diameter is ∼50 μm and the diameter of the hollow channel located at the center of the needle is ∼6 μm. The spacing between two adjacent needles is 150 μm. Optical images of the backside hole array are shown in Fig. 5h, where the diameter of the reservoir hole is ∼32 μm, and the spacing is 150 μm. The final reservoir diameter increased to 32 μm (it was originally 20 μm in TNT_type_I.gds at https://doi.org/10.6084/m9.figshare.16528311) because of SU8 shrinkage during the hard-bake at 250 °C86 (Step 1A(xiii)), but this widening is acceptable because the purpose is to store nucleic acid cargo. With ∼210-μm-long needles, the Type I chip can transport cargo more efficiently because of close contact with the skin. Figure 1c shows the cross-section view of the fabricated Type I needle array, after SiO2 PECVD coating, where the backside hollow bores are well aligned with the front-side needles. The cross section of one single needle, after cutting with a focused ion beam, is provided in Fig. 1d, showing the hollow bore being used as a pathway for plasmid DNA delivery.
Type II and Type III needles are expected to achieve a similar delivery result as Type I needles, but with additional functionality because of the tip sharpness. Figure 1i shows high-quality Type II silicon hollow-needle arrays with a sharp tip. The sharpness of the Type II needle arrays reduced the insertion force into tissue, allowing deeper penetration and delivery, and thus improved performance. A typical Type II needle has a length of ∼210 μm and similar diameter and bore size to those shown for Type I needles (∼6 μm) but has a sharp tip at the center. Figure 1j shows Type III silicon hollow-needle arrays with sharp tip and off-center bore. The tip is sharpened with the isotropic etching step (Step 1B(xviii)), generating a tip with a diameter as small as ∼12 μm. The hollow bore is designed with a deviation of 15 μm from the center of the needle to prevent tissue clogging on insertion.
In vivo TNT application
Electroporation-based methods have been reported to enhance the efficiency of plasmid penetration into target cells and tissues47,48,87. Using nanofabricated chips as outlined in this protocol, one can topically transfect tissue in seconds with minimal side effects. Here, we show that the depth of penetration of the fluorescently labeled FAM-DNA can be controlled through tuning the voltage of the electrical pulse (Fig. 12), and the area transfected is localized to the tissue directly under the chip. This method has been proven to be able to transfect multiple plasmids (see Reagents) and induce cell reprogramming in mouse skin3.
Fig. 12 |. Depth of cutaneous gene delivery as a function of applied voltage.
Representative fluorescent staining of dorsal murine skin after TNT to deliver FAM DNA (green) with Type I hollow-needle array at different voltages (50–200 V). The tissue sections were counter-stained with DAPI (blue). The sections were imaged using an AxioScan Z.1 (Zeiss) microscope.
The diagram in Fig. 11a and photo in Fig. 11b show the setup for TNT application in vivo. We used three plasmids encoding Ascl1, Brn2 and Myt1l (ABM) at a concentration of 0.5–0.1 μg/μl for TNT into mouse skin. Our ABM plasmid cocktail has been well characterized for its ability to convert fibroblasts to neuron-like cells77,88. For a control, we used an empty plasmid at the same total concentration. The skin was collected 24 h after transfection to quantify expression of the plasmids. We used RT-qPCR to quantify gene expression of both skin that underwent TNT with ABM and mock plasmid. After successful TNT, expression was significantly increased for all three ABM genes as shown in Fig. 11c. In vivo reprogramming of cells is highly dependent on the genes used for transfection. To evaluate reprogramming, we harvested skin for immunofluorescence 4 weeks after TNT. We evaluated the presence of the neuronal marker Tuj1+ and looked for Tuj1+fibers extending from a nucleus, because neuron cell bodies do not exist in normal skin, as shown in Fig. 11d.
Data availability
The individual images used to compile the figures shown and additional examples of the types of results obtained are available from Yi Xuan upon reasonable request. The design lithography patterns we used for needle fabrication are available at https://doi.org/10.6084/m9.figshare.16528311. Source data are provided with this paper.
Supplementary Material
Acknowledgements
This work made use of the Pritzker Nanofabrication Facility, which receives partial support from the SHyNE Resource, a node of the National Science Foundation’s National Nanotechnology Coordinated Infrastructure (NSF ECCS-2025633). We thank Parker Evans for his help in measuring the force applied during the TNT process. This work was supported in part by NIH grant DK128845; Department of Defense grants W81XWH-21-1-0097, W81XWH-21-1-0033 and W81XWH-20-1-251 to C.K.S.; Department of Defense grant W81XWH-21-1-0047 to S.K.; and NIH grant GM143572 to Y.X.
Peer review information Nature Protocols thanks Tarun Bhattacharyya, Kui Cheng, Yuanyu Huang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41596-021-00631-0. Correspondence and requests for materials should be addressed to Yi Xuan or Chandan K. Sen.
References
- 1.Abbasi J Nanochip turns skin into a bioreactor. JAMA 318, 898 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Miller MA Nanotransfection brings progress that’s more than skin-deep. Sci. Transl. Med. 9, eaao4216 (2017). [Google Scholar]
- 3.Gallego-Perez D et al. Topical tissue nano-transfection mediates non-viral stroma reprogramming and rescue. Nat. Nanotechnol. 12, 974–979 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zakrewsky M, Kumar S & Mitragotri S Nucleic acid delivery into skin for the treatment of skin disease:proofs-of-concept, potential impact, and remaining challenges. J. Control. Release 219, 445–456 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharei A et al. A vector-free microfluidic platform for intracellular delivery. Proc. Natl Acad. Sci. USA 110, 2082–2087 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y et al. Poking cells for efficient vector-free intracellular delivery. Nat. Commun. 5, 4466 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Pylaev T, Vanzha E, Avdeeva E, Khlebtsov B & Khlebtsov N A novel cell transfection platform based onlaser optoporation mediated by Au nanostar layers. J. Biophotonics 12, e201800166 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Xiong RH et al. Comparison of gold nanoparticle mediated photoporation: vapor nanobubbles outperformdirect heating for delivering macromolecules in live cells. ACS Nano 8, 6288–6296 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Boukany PE et al. Nanochannel electroporation delivers precise amounts of biomolecules into living cells. Nat. Nanotechnol. 6, 747–754 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Shi J et al. A review on electroporation-based intracellular delivery. Molecules 23, 3044 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kay MA, Glorioso JC & Naldini L Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nat. Med. 7, 33–40 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Knight S, Collins M & Takeuchi Y Insertional mutagenesis by retroviral vectors: current concepts and methods of analysis. Curr. Gene Ther. 13, 211–227 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Sawada S et al. Nanogel hybrid assembly for exosome intracellular delivery: effects on endocytosis and fusion by exosome surface polymer engineering. Biomater. Sci. 8, 619–630 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Yim N et al. Exosome engineering for efficient intracellular delivery of soluble proteins using optically reversible protein-protein interaction module. Nat. Commun. 7, 12277 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maas SLN, Breakefield XO & Weaver AM Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol. 27, 172–188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang QY et al. ARMMs as a versatile platform for intracellular delivery of macromolecules. Nat. Commun. 9, 960 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du JJ, Jin J, Yan M & Lu YF Synthetic nanocarriers for intracellular protein delivery. Curr. Drug Metab. 13, 82–92 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Cao Y et al. Nontoxic nanopore electroporation for effective intracellular delivery of biological macro-molecules. Proc. Natl Acad. Sci. USA 116, 7899–7904 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallego-Perez D et al. Deterministic transfection drives efficient nonviral reprogramming and uncovers reprogramming barriers. Nanomedicine 12, 399–409 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy S et al. Neurogenic tissue nanotransfection in the management of cutaneous diabetic polyneuropathy. Nanomedicine 128, 102220 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang D et al. Efficient delivery of nucleic acid molecules into skin by combined use of microneedle roller and flexible interdigitated electroporation array. Theranostics 8, 2361–2376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petchsangsai M, Rojanarata T, Opanasopit P & Ngawhirunpat T The combination of microneedles with electroporation and sonophoresis to enhance hydrophilic macromolecule skin penetration. Biol. Pharm. Bull. 37, 1373–1382 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Vinayakumar KB et al. A hollow stainless steel microneedle array to deliver insulin to a diabetic rat. bMicromech. Microeng. 26, 065013 (2016). [Google Scholar]
- 24.McAllister DV et al. Microfabricated needles for transdermal delivery of macromolecules and nanoparticles: fabrication methods and transport studies. Proc. Natl Acad. Sci. USA 100, 13755–13760 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller PR et al. Integrated carbon fiber electrodes within hollow polymer microneedles for transdermal electrochemical sensing. Biomicrofluidics 5, 13415 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mishra R, Maiti TK & Bhattacharyya TK Development of SU-8 hollow microneedles on a silicon substrate with microfluidic interconnects for transdermal drug delivery. J Micromech Microeng 28, 10.1088/1361-6439/aad301 (2018). [DOI] [Google Scholar]
- 27.Mishra R, Pramanick B, Maiti TK & Bhatracharyya TK Glassy carbon microneedles—new transdermal drug delivery device derived from a scalable C-MEMS process. Microsyst. Nanoeng. 4, 38 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gardeniers HJGE et al. Silicon micromachined hollow microneedles for transdermal liquid transport. Microelectromech. Syst. 12, 855–862 (2003). [Google Scholar]
- 29.Li Y et al. Fabrication of sharp silicon hollow microneedles by deep-reactive ion etching towards minimally invasive diagnostics. Microsyst. Nanoeng. 5, 41 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashrf M et al. Design, simulation and fabrication of silicon microneedles for bio-medical applications. Trans. Electr. Eng. Electron. Commun. 9, 83–91 (2011). [Google Scholar]
- 31.Wilke N, Mulcahy A, Ye SR & Morrissey A Process optimization and characterization of silicon microneedles fabricated by wet etch technology. Microelectron. J. 36, 650–656 (2005). [Google Scholar]
- 32.Kang SK et al. Bioresorbable silicon electronic sensors for the brain. Nature 530, 71–76 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Marty F et al. Advanced etching of silicon based on deep reactive ion etching for silicon high aspect ratio microstructures and three-dimensional micro- and nanostructures. Microelectron. J. 36, 673–677 (2005). [Google Scholar]
- 34.Ji J, Tay FEH, Miao JM & Iliescu C Microfabricated silicon microneedle array for transdermal drug delivery. J. Phys. Conf. Ser. 34, 1127–1131 (2006). [Google Scholar]
- 35.Wilke N, Hibert C, O’Brien J & Morrissey A Silicon microneedle electrode array with temperature monitoring for electroporation. Sens. Actuat. A Phys. 123–124, 319–325 (2005). [Google Scholar]
- 36.Lai SL, Johnson D & Westerman R Aspect ratio dependent etching lag reduction in deep silicon etch processes. J. Vac. Sci. Technol. A 24, 1283–1288 (2006). [Google Scholar]
- 37.Tang Y, Sandoughsaz A, Owen KJ & Najafi K Ultra deep reactive ion etching of high aspect-ratio and thick silicon using a ramped-parameter process. J. Microelectromech. Syst. 27, 686–697 (2018). [Google Scholar]
- 38.Collins F Tissue nanotransfection: skin cells can be reprogrammed in vivo. https://directorsblog.nih.gov/2019/02/14/skin-cells-can-be-reprogrammed-in-vivo/ (NIHDirector’sBlog,2019).
- 39.Zhou X et al. Exosome-mediated crosstalk between keratinocytes and macrophages in cutaneous wound healing. ACS Nano 14, 12732–12748 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore JT et al. Nanochannel-based poration drives benign and effective nonviral gene delivery to peripheral nerve tissue. Adv. Biosyst. 4, e2000157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cunningham JJ, Ulbright TM, Pera MF & Looijenga LH Lessons from human teratomas to guide development of safe stem cell therapies. Nat. Biotechnol. 30, 849–857 (2012); erratum 31, 565 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Losordo DW & Dimmeler S Therapeutic angiogenesis and vasculogenesis for ischemic disease: part II: cell-based therapies. Circulation 109, 2692–2697 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Mount NM, Ward SJ, Kefalas P & Hyllner J Cell-based therapy technology classifications and translational challenges. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20150017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luckay A et al. Effect of plasmid DNA vaccine design and in vivo electroporation on the resulting vaccine specific immune responses in rhesus macaques. J. Virol. 81, 5257–5269 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vargas JE et al. Retroviral vectors and transposons for stable gene therapy: advances, current challenges and perspectives. J. Transl. Med. 14, 288 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aihara H & Miyazaki J -i Gene transfer into muscle by electroporation in vivo. Nat. Biotechnol. 16, 867–870 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Mir LM et al. High-efficiency gene transfer into skeletal muscle mediated by electric pulses. Proc. Natl Acad. Sci. USA 96, 4262–4267 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin F et al. Optimization of electroporation-enhanced intradermal delivery of DNA vaccine using a minimally invasive surface device. Hum. Gene Ther. Methods 23, 157–168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matriano JA et al. Macroflux microprojection array patch technology: a new and efficient approach for intracutaneous immunization. Pharm. Res. 19, 63–70 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Daugimont L et al. Hollow microneedle arrays for intradermal drug delivery and DNA electroporation. J. Membr. Biol. 236, 117–125 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Park JH, Allen MG & Prausnitz MR Biodegradable polymer microneedles: fabrication, mechanics and transdermal drug delivery. J. Control. Release 104, 51–66 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Park JH, Yoon YK, Choi SO, Prausnitz MR & Allen MG Tapered conical polymer microneedles fabricated using an integrated lens technique for transdermal drug delivery. IEEE Trans. Biomed. Eng. 54, 903–913 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Sullivan SP et al. Dissolving polymer microneedle patches for influenza vaccination. Nat. Med. 16, 915–920 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Maaden K et al. Hollow microneedle-mediated micro-injections of a liposomal HPV E7(43–63) synthetic long peptide vaccine for efficient induction of cytotoxic and T-helper responses. J. Control. Release 269, 347–354 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Kim YC, Park JH & Prausnitz MR Microneedles for drug and vaccine delivery. Adv. Drug Deliv. Rev. 64, 1547–1568 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Narayanan SP & Raghavan S Solid silicon microneedles for drug delivery applications. Int. J. Adv. Manuf. Tech. 93, 407–422 (2017). [Google Scholar]
- 57.Xie X et al. Nanostraw–electroporation system for highly efficient intracellular delivery and transfection. ACS Nano 7, 4351–4358 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Cao Y et al. Nondestructive nanostraw intracellular sampling for longitudinal cell monitoring. Proc. Natl Acad. Sci. USA 114, E1866–E1874 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He G et al. Fabrication of various structures of nanostraw arrays and their applications in gene delivery. Adv. Mater. Interfaces 5, 1701535 (2018). [Google Scholar]
- 60.He G et al. Multifunctional branched nanostraw-electroporation platform for intracellular regulation and monitoring of circulating tumor cells. Nano Lett. 19, 7201–7209 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Tay A & Melosh N Nanostructured materials for intracellular cargo delivery. Acc. Chem. Res. 52, 2462–2471 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wen R et al. Intracellular delivery and sensing system based on electroplated conductive nanostraw arrays. ACS Appl. Mater. Interfaces 11, 43936–43948 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Gill HS & Prausnitz MR Coated microneedles for transdermal delivery. J. Control. Release 117, 227–237 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DeMuth PC, Su X, Samuel RE, Hammond PT & Irvine DJ Nano-layered microneedles for transcutaneous delivery of polymer nanoparticles and plasmid DNA. Adv. Mater. 22, 4851–4856 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim H et al. Bioresorbable, miniaturized porous silicon needles on a flexible water-soluble backing for unobtrusive, sustained delivery of chemotherapy. ACS Nano 14, 7227–7236 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chiappini C et al. Biodegradable silicon nanoneedles delivering nucleic acids intracellularly induce localizedin vivo neovascularization. Nat. Mater. 14, 532–539 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang B, Shi Y, Miyamoto D, Nakazawa K & Miyake T Nanostraw membrane stamping for direct delivery of molecules into adhesive cells. Sci. Rep. 9, 6806 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seong H et al. Size-tunable nanoneedle arrays for influencing stem cell morphology, gene expression, and nuclear membrane curvature. ACS Nano 14, 5371–5381 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen W, Li H, Shi D, Liu Z & Yuan W Microneedles as a delivery system for gene therapy. Front. Pharmacol. 7, 137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dul M et al. Hydrodynamic gene delivery in human skin using a hollow microneedle device. J. Control. Release 265, 120–131 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Bolhassani A, Khavari A & Orafa Z Electroporation—advantages and drawbacks for delivery of drug, gene and vaccine. in Application of Nanotechnology in Drug Delivery (InTech, 2014). [Google Scholar]
- 72.Huo Z-Y et al. Carbon-nanotube sponges enabling highly efficient and reliable cell inactivation by low-voltage electroporation. Environ. Sci. Nano 4, 2010–2017 (2017). [Google Scholar]
- 73.Hyder I, Eghbalsaied S & Kues WA Systematic optimization of square-wave electroporation conditions for bovine primary fibroblasts. BMC Mol. Cell Biol. 21, 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu Y, Werner C & Li D Electrokinetic transport through rough microchannels. Anal. Chem. 75, 5747–5758 (2003). [DOI] [PubMed] [Google Scholar]
- 75.Fu J et al. Improving sidewall roughness by combined RIE-Bosch process. Mat. Sci. Semicon. Proc. 83, 186–191 (2018). [Google Scholar]
- 76.Chutani RK, Hasegawa M, Maurice V, Passilly N & Gorecki C Single-step deep reactive ion etching ofultra-deep silicon cavities with smooth sidewalls. Sens. Actuators A Phys. 208, 66 (2014). [Google Scholar]
- 77.Canatella PJ, Karr JF, Petros JA & Prausnitz MR Quantitative study of electroporation-mediated molecular uptake and cell viability. Biophys. J. 80, 755–764 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stewart MP et al. In vitro and ex vivo strategies for intracellular delivery. Nature 538, 183–192 (2016). [DOI] [PubMed] [Google Scholar]
- 79.Fei Z et al. Micronozzle array enhanced sandwich electroporation of embryonic stem cells. Anal. Chem. 82, 353–358 (2010). [DOI] [PubMed] [Google Scholar]
- 80.Chang L et al. Magnetic tweezers-based 3D microchannel electroporation for high-throughput gene transfection in living cells. Small 11, 1818–1828 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cao Y et al. Reply to Nathamgari et al.: nanopore electroporation for intracellular delivery of biological macromolecules. Proc. Natl Acad. Sci. USA 116, 22911 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Herrick A, Perry AJ & Boswell RW Etching silicon by SF6 in a continuous and pulsed power helicon reactor. J. Vac. Sci. Technol. A 21, 955–966 (2003). [Google Scholar]
- 83.Wongwanitwattana C et al. Precision plasma etching of Si, Ge, and Ge:P by SF6 with added O2. J. Vac. Sci. Technol. A 32, 031302 (2014). [Google Scholar]
- 84.Shikida M, Hasada T & Sato K Fabrication of a hollow needle structure by dicing, wet etching and metal deposition. J. Micromech. Microeng. 16, 2230–2239 (2006). [Google Scholar]
- 85.Yan G, Warner KS, Zhang J, Sharma S & Gale BK Evaluation needle length and density ofmicroneedle arrays in the pretreatment of skin for transdermal drug delivery. Int. J. Pharmaceutics 391, 7–12 (2010). [DOI] [PubMed] [Google Scholar]
- 86.Natu R, Islam M, Gilmore J & Martinez-Duarte R Shrinkage of SU-8 microstructures during carbonization. J. Anal. Appl. Pyrolysis 131, 17–27 (2018). [Google Scholar]
- 87.Miyazaki J.-i. & Aihara H Gene transfer into muscle by electroporation in vivo. in Gene Therapy Protocols 2nd edn (ed. Morgan JR). 49–62 (Springer, 2002). [DOI] [PubMed] [Google Scholar]
- 88.Zhang X et al. Characteristics of liquid flow in microchannels at very low Reynolds numbers. Chem. Eng. Technol. 39, 1425–1430 (2016). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The individual images used to compile the figures shown and additional examples of the types of results obtained are available from Yi Xuan upon reasonable request. The design lithography patterns we used for needle fabrication are available at https://doi.org/10.6084/m9.figshare.16528311. Source data are provided with this paper.