FIGURE 1.
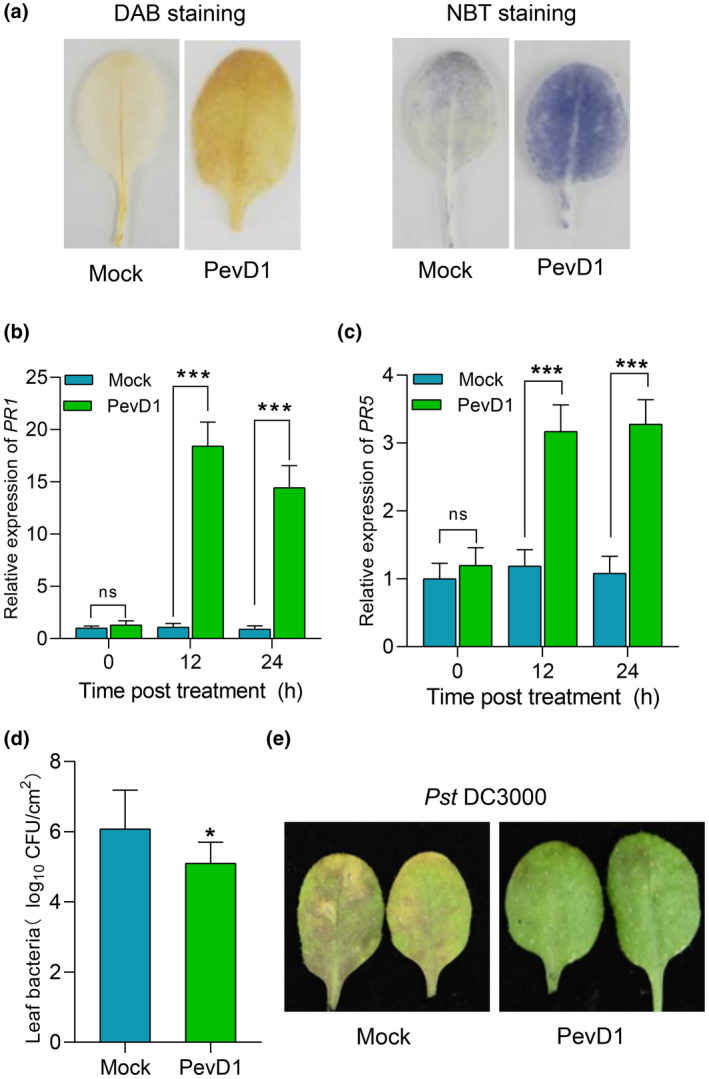
PevD1 triggers a defence response and enhances disease resistance to Pseudomonas syringae pv. tomato (Pst) DC3000 in Arabidopsis thaliana (Col‐0). (a) Photographs of 3,3′‐diaminobenzidine (DAB)‐ and nitroblue tetrazolium (NBT)‐stained leaves infiltrated with buffer (Mock) or PevD1. Four‐week‐old wild‐type Col‐0 leaves were infiltrated with PevD1 or Tris buffer (Mock) and stained with DAB and NBT at 24 h postinfiltration (hpi). (b,c) Relative expression levels of PR1 and PR5 in 4‐week‐old wild‐type Col‐0 leaves at 0, 12, and 24 hpi. UBC21 was used as the internal control, the PR1 and PR5 expression is represented relative to the UBC21 transcription level. The bars were calculated based on three independent experiments. The values are mean ± SD (n = 3). ***p < 0.001, one‐way analysis of variance (ANOVA). (d) Leaf bacteria was measured by colony counting. Col‐0 leaves were infiltrated with PevD1 or Tris buffer (Mock) and inoculated with Pst DC3000 at 24 hpi. Bacterial colonies were counted at 48 hpi. Data from three separate experiments are shown (mean ± SD, n = 6). *p < 0.05, one‐way ANOVA. (e) Photographs of Pst DC3000‐infected disease symptoms in leaves. Col‐0 leaves were infiltrated with PevD1 or Tris buffer (Mock) then inoculated with Pst DC3000 at 24 hpi. Photograph was taken at 48 hpi
