FIGURE 2.
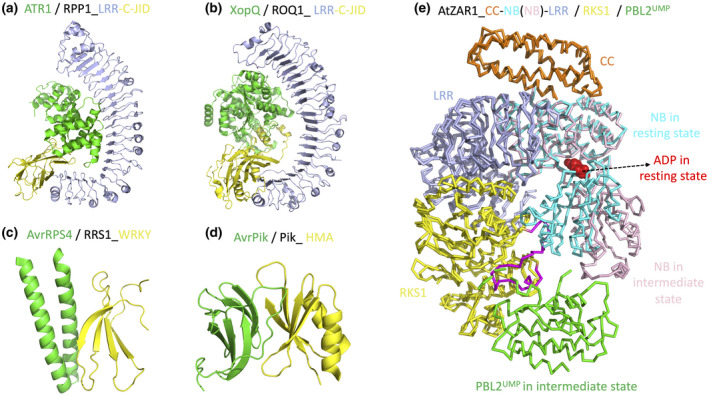
Current structure knowledge on NLR recognition of effectors. (a) Cryoelectron microscopy (cryo‐EM) structure of RPP1 LRR and C‐JID domains in a complex with effector ATR1 (PDB: 7CRB). (b) Cryo‐EM structure of ROQ1 LRR and C‐JID domains in a complex with effector XopQ (PDB: 7JLU). (c) Crystal structure of RRS1 WRKY domain in a complex with effector AvrRPS4 (PDB: 7P8K). (d) Crystal structure of Pik HMA domain in a complex with effector AvrPik (PDB: 5A6W). (e) Super‐imposition of the ADP‐bound ZAR1 structure in the resting state (6J5W) and the ADP‐free ZAR1 structure in the intermediate state (6J5V). Binding of effector‐produced PBL2UMP causes RKS1 conformational changes (highlighted in purple) that clash with and push away the NB domain. The NB domain is shown in two colours (cyan and pink) to highlight the close‐to‐open switch that allows allosteric ADP release
