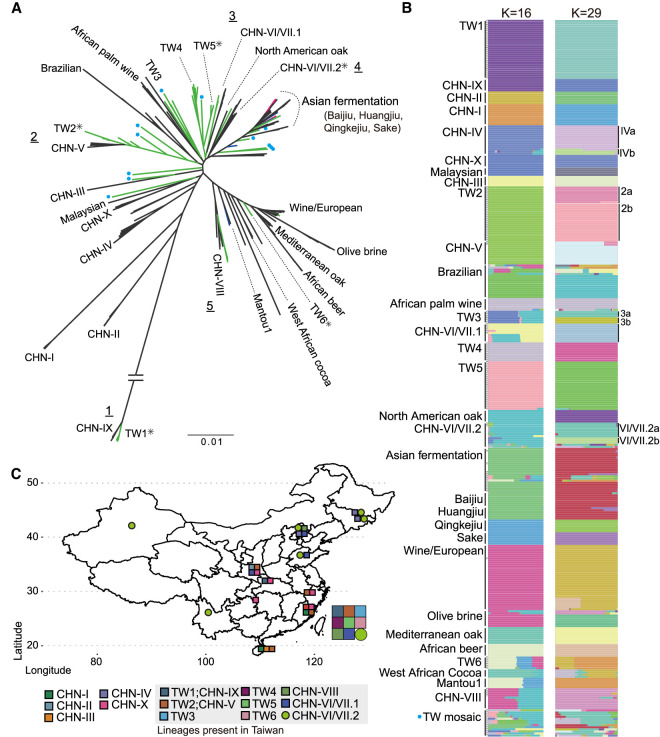
Figure 2.
Phylogeny and population structures of 340 S. cerevisiae isolates. (A) Unrooted phylogeny based on 765,169 genome-wide SNPs. Bootstrap support was >90% in all major lineages except inner nodes within some lineages, as indicated by asterisks. Natural, industrial, and fermentation-related isolates discovered in Taiwan are colored in green, blue, and magenta, respectively. Mosaic Taiwanese isolates from ADMIXTURE analyses are labeled with blue dots on branch tips. Five cases in which Taiwanese and Chinese isolates were found to be monophyletic are indicated with underscored numbers. The Asian fermentation lineage includes Baijiu-, Huangjiu-, Qingke jiu-, sake-, and fermentation-related isolates from Taiwan, as shown in B. (B) Population structure from ADMIXTURE analysis at K = 16 and 29. Labels on the left side of the bars indicate each group from K = 16, and some were further separated in K = 29, which is annotated on the right side. Natural Taiwanese isolates with admixed genome makeup are shown together in the TW mosaic group. (C) Map of China and Taiwan indicating where the S. cerevisiae natural lineages were found (colored squares and circle). CHN-IV isolates that were sampled from Japan are not shown on this map.