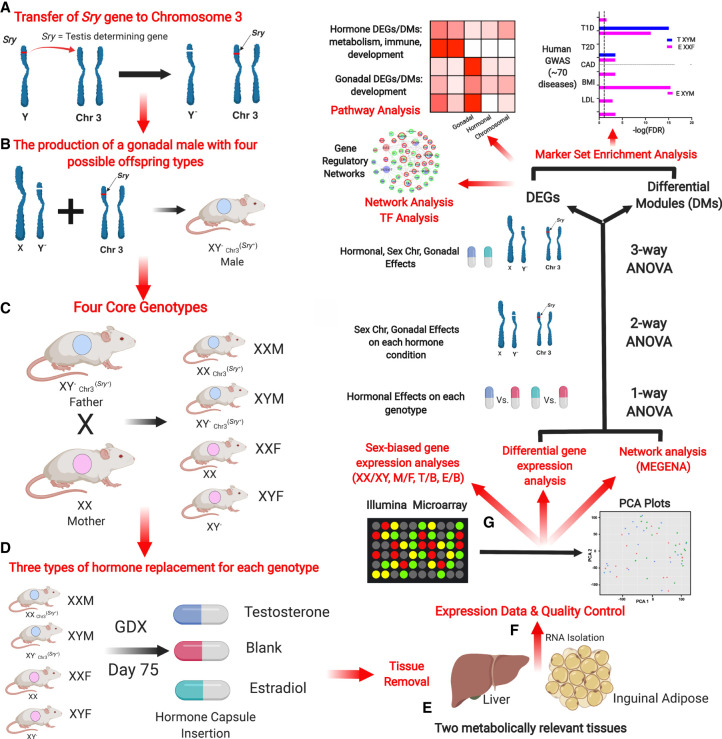
Figure 1.
Overall study design. (A) Transfer of the Sry gene to Chromosome 3. Sry, which is usually located on the Y Chromosome, was deleted (a spontaneous deletion) and inserted as a transgene onto Chromosome 3, making Sry independent of the Y Chromosome. (B) The production of a gonadal male XY− Chr3Sry+, which has the ability to produce four types of gametes resulting in the four core genotypes (FCG). (C) The generation of the FCG mice. Mating of XY− Sry+ male and XX female produces four types of mouse offspring (two gonadal males and two gonadal females): XY−Chr3Sry+ (XYM), XXSry+ (XXM), XX (XXF), XY− (XYF). (D) Modulation of sex hormones in mouse offspring of each genotype after gonadectomy (GDX). Each of the four core genotypes underwent GDX at day 75 and was implanted with a capsule that contained either estradiol or testosterone, or was blank (n = 5/genotype/treatment). (E) Dissection of the liver and inguinal adipose tissue for RNA isolation. (F) Gene expression profiling and quality control. Using an Illumina microarray, we measured the transcriptome and then performed a principal component analysis (PCA) to identify outliers and global patterns. (G) Bioinformatics analyses. Differentially expressed genes (DEGs) influenced by individual sex-biasing factors were identified using three-way ANOVA (chromosomal, gonadal, and hormonal effects), two-way ANOVA (gonadal and chromosomal effects under each hormone condition), and a one-way ANOVA (estradiol and testosterone treatment effects in individual genotypes). Gene coexpression networks were constructed using MEGENA and differential coexpression modules (DMs) affected by individual sex-biasing factors were identified using three-way, two-way, and one-way ANOVAs. DEGs and DMs were analyzed for enrichment of functional categories or biological pathways. The relevance of the DEGs to human disease was assessed via integration with human genome-wide association studies (GWAS) for more than 70 diseases using the marker set enrichment analysis (MSEA). Transcription factor analysis and gene regulatory network analysis were additionally conducted on the DEGs derived from the one-way ANOVA.