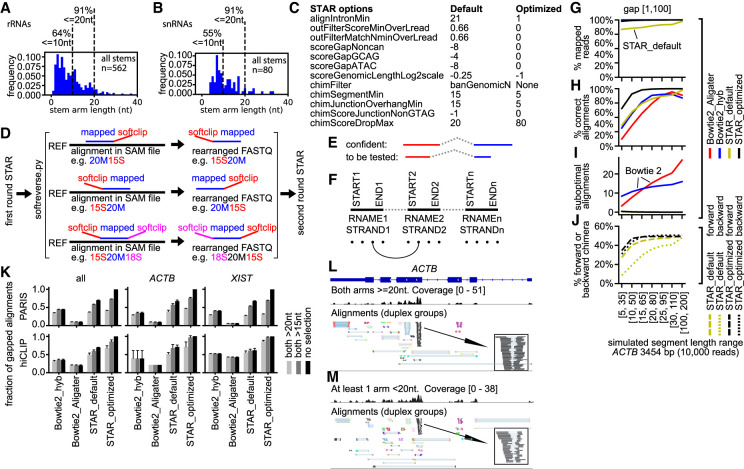
Figure 2.
Optimization of short-read mapping from crosslink-ligation experiments. (A,B) RNA stems were extracted from the human cytoplasmic and mitochondrial ribosome and spliceosome crystal or cryo-EM structures. The following RNAs are included: 12S, 16S, 5S, 5.8S, 18S, 28S, U1, U2, U4, U6, U5, U11, U12, U4atac, and U6atac. (C) List of critical STAR parameters that are optimized to map noncontinuous reads. The default value for chimSegmentMin is unset, whereas setting this value to any positive integer triggers chimeric alignments. The recommended value of 15 is used here as the “default.” (D) Strategy for the two-round STAR mapping. After the first round of optimized STAR mapping, continuous alignments with softclips (“S” in CIGAR) are rearranged and then mapped again using the optimized STAR parameters. (E,F) Strategies for filtering alignments after STAR mapping. (E) Confident alignments: all segments or arms are uniquely mapped to the genome. Alignments with shorter segments that cannot be mapped uniquely are to be tested against confident ones. (F) Filtering method for the less confident alignments: all arms of the confident alignments are built into a database of connections between segments, in five nucleotide intervals (dots shown at the bottom). The connection database consists of reference name (RNAME), strand (STRAND), and coordinates between start and end (START, END). Then, the less confident alignments are tested against this database. (G–J) Benchmarking four mapping strategies on simulated reads for the human ACTB gene. Alignments are quantified on the following four aspects: (G) % mapped reads, that is, reads that are mappable to hg38 primary genome; (H) % correct alignments, that is, alignments with the same mapped positions and gap lengths as the simulated values, allowing 10-nt differences in positions or lengths due to ambiguities at the ends of reads; (I) Suboptimal alignments per read, defined as alignments that are not mapped to the correct locations; (J) % forward or backward chimera. In theory, both forward and backward chimera should be ∼50% (randomly assigned during simulation, so they are not precisely 50%). Here, only STAR alignments are calculated. (K) Gap1 (one gap, i.e., two segments) alignments in PARIS and hiCLIP data were recovered by various mapping methods and segment-length selections. Fractions for the highest-performing method (STAR_optimized) are set to 1. For STAR analysis, sequencing reads were mapped to the genome (hg38 primary); then alignments were filtered and classified into six categories using gaptypes.py. The gap1 alignments were filtered to remove short gaps and splicing alignments (gapfilter.py). Primary alignments were extracted from all alignments and used for analysis. For Bowtie 2 mapping, previously reported parameters (hyb and Aligater) were used. Unique alignments with deletions (D in SAM CIGAR string) were extracted and alignments were converted to join the multiple segments (bowtie2chim.py). Then, the alignments were classified using gaptypes.py. The gap1 alignments were filtered to remove short gaps and splicing alignments (gapfilter.py). The selection of alignments with both arms > 15 nt or 20 nt mimics the mapping and chaining strategy in previous studies that employ Bowtie 2 (hyb and Aligater). (L,M) Alignments in the ACTB mRNA from PARIS data in HEK cells were separated into ones where both arms (or segments) are at least 20 nt (L), or at least one arm is shorter than 20 nt (M). The inset boxes show DGs that support the same duplex regardless of segment length.