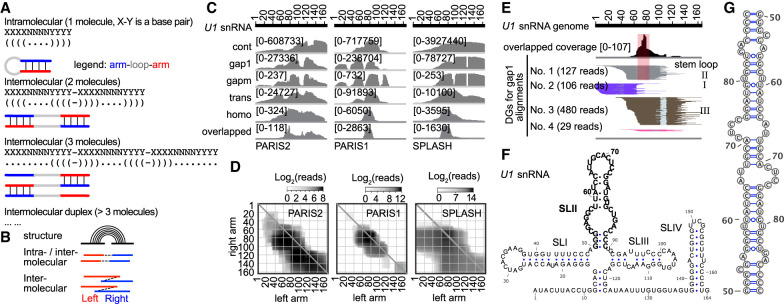
Figure 6.
Identification of potential RNA homodimers using homotypic alignments. (A) The same base-pairing interactions can mediate intramolecular stem–loops (top) and homotypic interactions between two (middle) or more (bottom) copies of the same molecule. (B) Diagram showing alignments with gapped or overlapped arms, suggesting RNA stem–loops or homodimers. (C) Coverage of five different types of alignments on U1. The overlapped part of homo alignments is shown individually at the bottom. (D) Heat map of U1 snRNA homo alignments in three data sets. (E) PARIS2 data showing overlapped regions and corresponding local stem–loop (SLII). DGs were assembled from 1000 total alignments. (F) Secondary structure of U1homo interaction, with the SLII in bold letters. (G) Secondary structure model for the SLII homodimer.