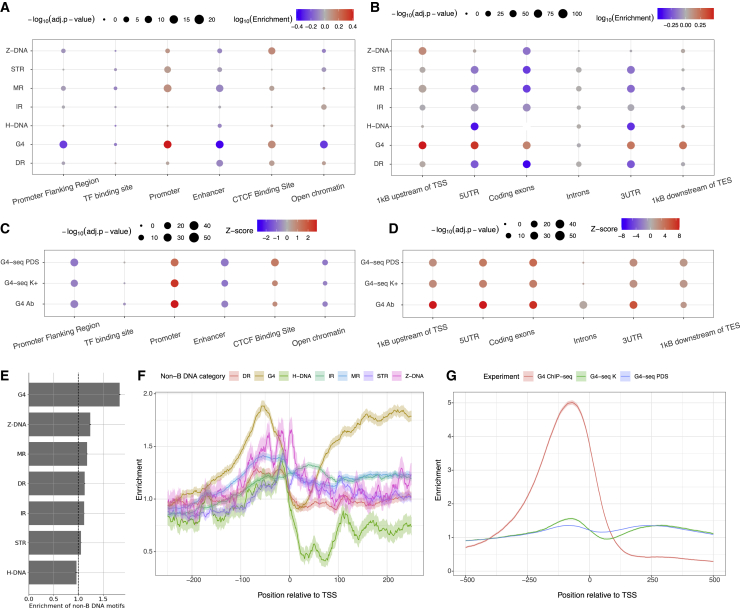
Figure 2.
Non-B DNA motifs at functional elements
(A) Median relative enrichment across 12 cell lines for non-B DNA motif enrichment at Ensembl Regulatory Features.
(B) Non-B DNA motif enrichment at functional genomic compartments for each non-B DNA motif. Statistical significance was estimated using Binomial tests with Bonferroni correction.
(C) Z score of G4-seq and G4 ChIP-seq peak density across Ensembl Regulatory Features.
(D) Z score of G4-seq and G4 ChIP-seq peak density across the gene body.
For (C) and (D), two treatments that stabilize G4s, PDS and K+, were used in G4-seq.
(E) Enrichment of non-B DNA motifs in the [–250, 0] region relative to the wider promoter region (–1 kB, 0). Error bars represent standard deviation from bootstrapping.
(F) Base-pair resolution of distribution of nucleotide motifs relative to the TSS. IRs, MRs, DRs, STRs, and G4s are abbreviations for inverted repeats, mirror repeats, direct repeats, short tandem repeats, and G-quadruplexes, respectively.
(G) G4 enrichment patterns relative to the TSS for G4 motif, G4-seq peaks in K+ and PDS treatments, and from G4 ChIP-seq peaks.