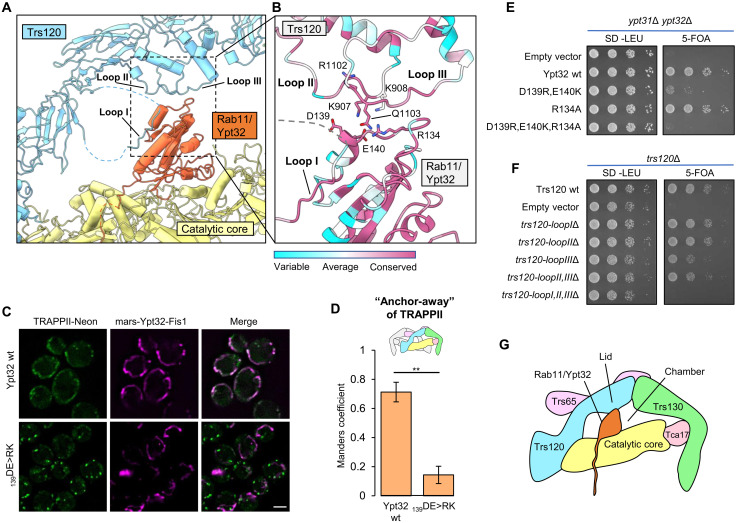
Fig. 3. Trs120 forms a lid required for Rab11/Ypt32 interaction.
(A) View of the Rab11/Ypt32 binding interface with Trs120. Ordered loops that contact Rab11/Ypt32 are indicated. (B) Close-up view of (A), with residue conservation indicated. (C) Imaging data from a GRab-IT experiment monitoring the association between TRAPPII and nucleotide-free wild-type or mutant Rab11/Ypt32. Note that both “wild-type” and mutant Rab11/Ypt32 constructs harbor a mutation that prevents nucleotide binding (see Materials and Methods). (D) Quantitation of the data in (C). **P < 0.01. (E) Complementation test assessing the effects of residue substitutions in Rab11/Ypt32 on cell viability. 5-FOA, 5-fluoroorotic acid. (F) Complementation test assessing the effects of loop deletion mutants of Trs120 on cell viability. (G) Cartoon schematic illustrating the interaction of Rab11/Ypt32 with the TRAPPII monomer. Trs120 forms a lid to enclose Ypt32 within the active site chamber. Scale bar, 2 μm.