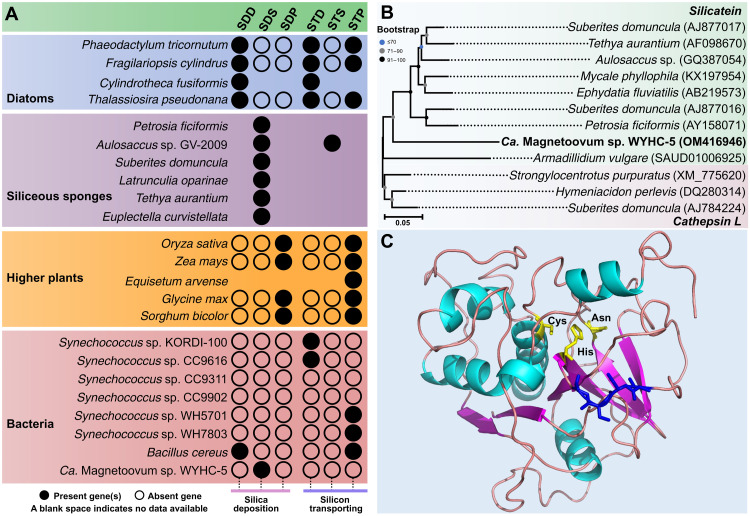
Fig. 4. Prediction of Si-related gene and protein in strain WYHC-5.
(A) Distribution of silica deposition (SD) and silicon transporting (ST) genes in typical silicifying eukaryotes and prokaryotes (including strain WYHC-5). The SDD, SDS, and SDP represent the silica deposition proteins identified in diatoms [including silaffin, silacidin, silicalemma-associated proteins (SAP), silicanin, and cingulin], siliceous sponges (including silicatein, glassin, and silintaphin), and higher plants (siliplant1). STD, STS, and STP represent the silicon-transporting proteins identified in diatoms [including SIT (silicon transporter) and SIT-like], siliceous sponges [Na+/HCO3− [Si(OH)4] co-transporter (NBCSA)] and higher plants (including lsi1, lsi2, lsi2-like, lsi3, lsi6, OsCASP1, NIP2, and NIP3). More detailed data are showed in data files S3 to S5. (B) Cluster analysis of the cDNA sequences of silicatein (light green background) in siliceous sponges, A. vulgare, strain WYHC-5, and cathepsin L (light red background) in siliceous sponges. Bootstrap values at nodes are given as percentages of 1000 replicates. GenBank accession numbers are given in parentheses. (C) Modeling of the 3D structure of the WYHC-5 silicatein-1. The GMQE and QMEAN scores are 0.71 and 0.69 ± 0.05, respectively, indicating that the model structure is accurate and shows good agreement with the template. The catalytic center of silicatein-1, comprising the amino acids Cys, His, and Asn (yellow), and the localization of the serine cluster (blue) are marked.