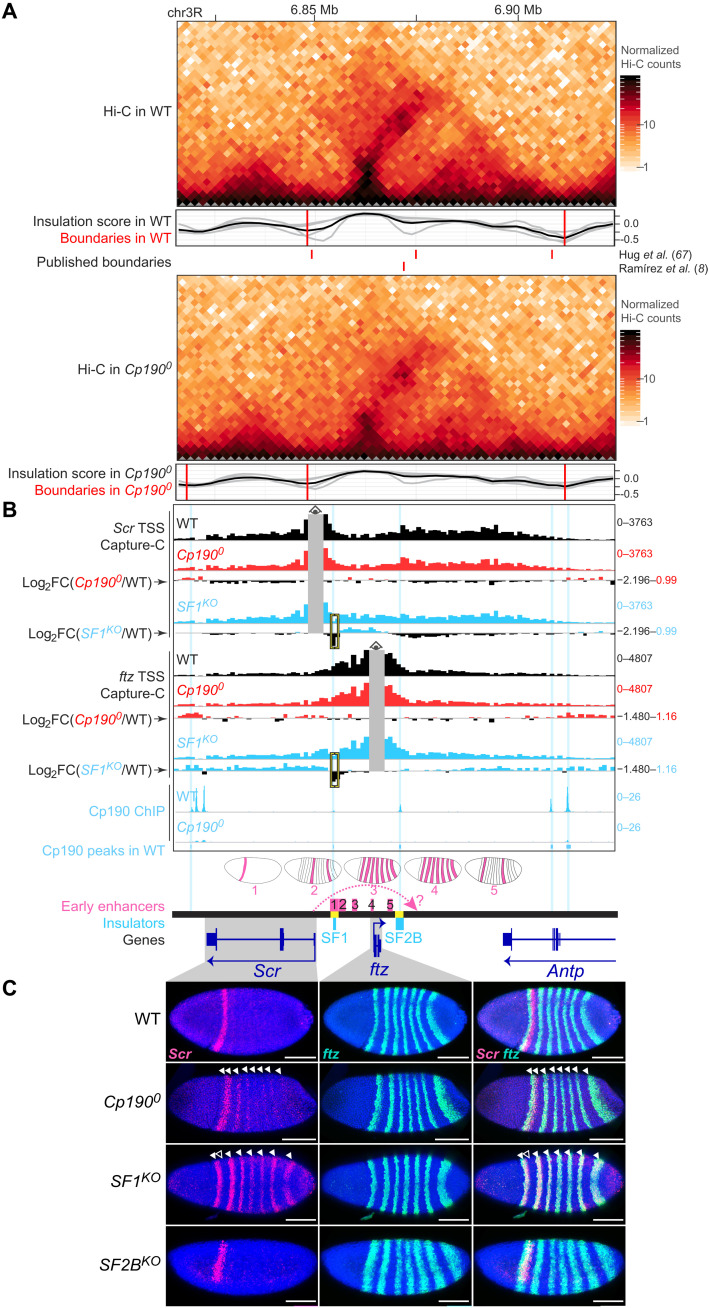
Fig. 5. Cp190 prevents regulatory cross-talk between early patterning gene loci.
(A) Extended ftz locus (dm6 coordinates) Hi-C maps presented as in Fig. 1E. (B) NG Capture-C profiles (1-kb resolution) around Scr or ftz TSS viewpoints showing average normalized reads (in reads per million) of biological triplicates excluding bins ±2 kb around the viewpoint (gray). Differential Capture-C profiles in mutant versus WT are shown as log2 fold change profiles obtained from diffHic. Yellow brackets mark the deleted boundary in SF1KO. WT Cp190 ChIP peaks are highlighted in blue (two prominent Cp190 ChIP signals in WT overlap a blacklisted region and were not called peaks). Early stripe enhancers with schematized expression patterns are numbered 1 to 5 (references in Materials and Methods). Dotted arrow shows Scr regulation by a hypothetical distal enhancer (question mark). SF1KO and SF2BKO deletions are yellow. (C) RNA–fluorescence in situ hybridization (FISH) with cohybridized antisense probes against Scr (red) and ftz (green) mRNAs in 4′,6-diamidino-2-phenylindole (DAPI)–stained early gastrula embryos (anterior left; posterior right; scale bars, 100 μm; merged images on the right). In Cp1900, Scr is expressed in its WT stripe and in ectopic ftz stripes (filled arrowheads). In SF1KO embryos, Scr is lost in its WT stripe (empty arrowhead) and only expressed in ftz stripes (filled arrowheads). In SF2BKO embryos, Scr and ftz expression seem normal (embryo rotation reveals a normal ventral gap in Scr expression).