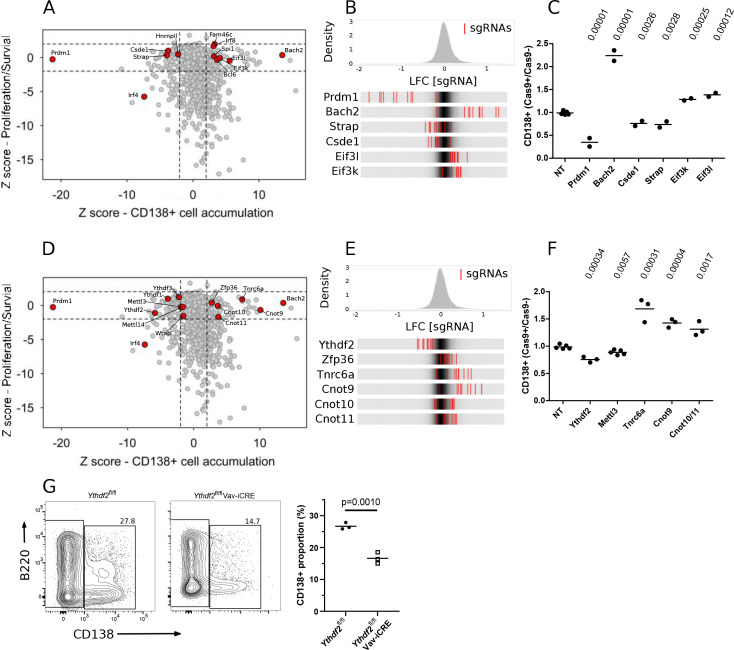
Figure 1. Genetic screen identifies modulators of B cell expansion and plasma cell accumulation.
(A) Dot plot representation of genetic screens of B cell proliferation/survival, and CD138 + cell accumulation: X-axis shows z-score of gene-level log2 fold change (LFC) for CD138 + cell accumulation (CD138 +v CD138- cells) and Y-axis shows z-score of gene-level LFC (day 8 v day 4 cells) for B cell expansion calculated by MAGECK. (B) Top: Distribution of enrichments of all sgRNAs (z-scores of sgRNA-level LFC: CD138 +v CD138- cells) for the CD138 + cell accumulation screen. Bottom: Enrichment for each of the ten sgRNAs, represented as red lines, targeting one of the indicated genes in comparison to the overall distribution of sgRNAs in the screen, depicted as the grey density in the middle of each bar. (C) The ratio of the proportion of cells expressing CD138 in Cas9 + cells and Cas9- cells transduced by viruses with non-targeting (NT), Prmd1, Bach2, Csde1, Strap, Eif3k, Eif3l targeting sgRNAs at day-8 of the in vitro B cell culture the data are representative of between two and four experiments performed on different days. Statistical significance was determined by two-tailed unpaired Student’s t-test, p values unadjusted for multiple testing are plotted. Each symbol is representative of a distinct sgRNA. (D) Same as in (A) with additional genes highlighted. (E) Same as in (B) with additional genes highlighted. (F) Same as in (C) with Ythdf2, Mettl3, Tnrc6a, Cnot9 targeting sgRNAs and paired sgRNAs against Cnot10/Cnot11. the data are representative of between two and four experiments performed on different days. (G) Left: representative flow cytometry for Ythdf2 CTL and Ythdf2 CKO and right: summary data of the proportion of cells expressing CD138 at day 8 of an in vitro culture of B cells from Ythdf2fl/fl mice (closed circles) or Ythdf2fl/fl-Vav1-iCre mice (open squares); the data are representative of three experiments performed on different days. Statistical significance was determined by two-tailed unpaired Student’s t-test. Each symbol is representative of cells from a single mouse.