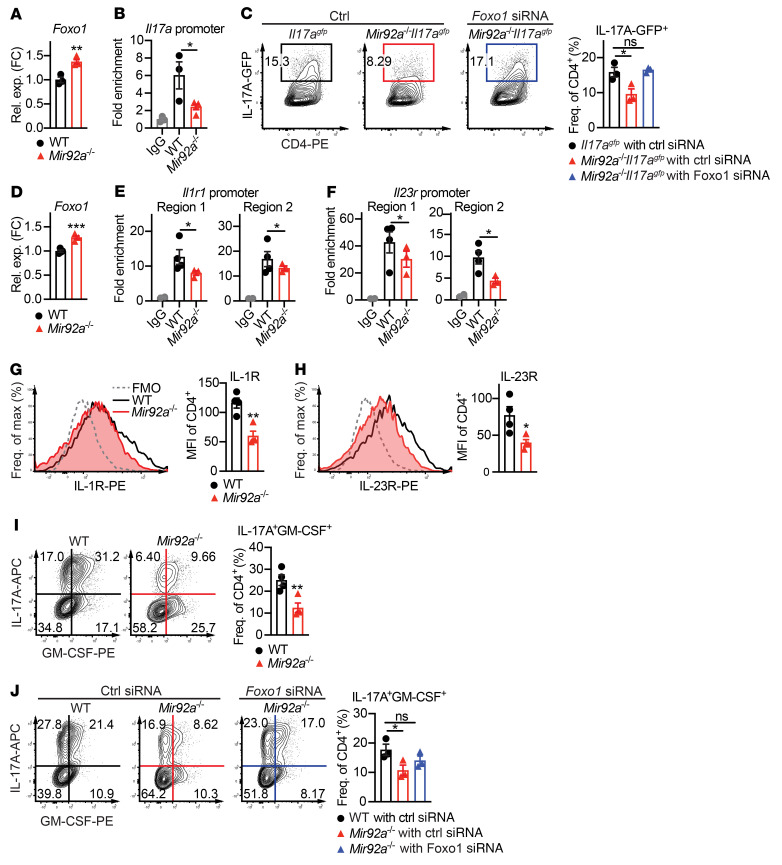
Figure 5. miR-92a promotes nonpathogenic and pathogenic Th17 development by targeting Foxo1.
(A) qPCR analysis of Foxo1 expression in nonpathogenic Th17-polarized WT and Mir92a–/– naive CD4+ T cells (n = 3–4). (B) ChIP analysis of RORγt binding to the Il17a locus in nonpathogenic Th17-polarized WT and Mir92a–/– naive CD4+ T cells (n = 3–4). (C) Representative flow cytometric plots and frequencies of IL-17A–GFP+ cells in Il17agfp and Mir92a–/– Il17agfp naive CD4+ T cells, transfected in vitro with control or Foxo1 siRNA and then cultured under nonpathogenic Th17-polarizing conditions (n = 3). (D) qPCR analysis of Foxo1 expression in pathogenic Th17-polarized WT and Mir92a–/– naive CD4+ T cells (n = 4). (E and F) ChIP analyses of RORγt binding to the Il1r1 promoter loci (E) and Il23r promoter loci (F) in pathogenic Th17-polarized WT and Mir92a–/– naive CD4+ T cells (n = 3–4). (G and H) Representative flow cytometric histograms and MFIs of IL-1R (G) and IL-23R (H) in pathogenic Th17-polarized WT and Mir92a–/– CD4+ T cells (n = 4). MFI values shown were obtained after subtracting the MFI values of fluorescence minus one (FMO) controls for IL-1R or IL-23R. (I) Representative flow cytometric plots and frequencies of IL-17A+GM-CSF+ cells from G and H (n = 4). (J) Representative flow cytometric plots and frequencies of IL-17A+GM-CSF+ cells in WT and Mir92a–/– naive CD4+ T cells transfected with control or Foxo1 siRNA followed by culturing under pathogenic Th17-polarizing conditions (n = 3). Fold enrichment is shown relative to WT IgG conditions (B, G, and H). Data are representative of 2 independent experiments and indicate the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, by unpaired, 2-tailed Student’s t test (A, D, G–I) or 1-way ANOVA with Dunnet’s multiple-comparison test (B, C, E, F, and J).