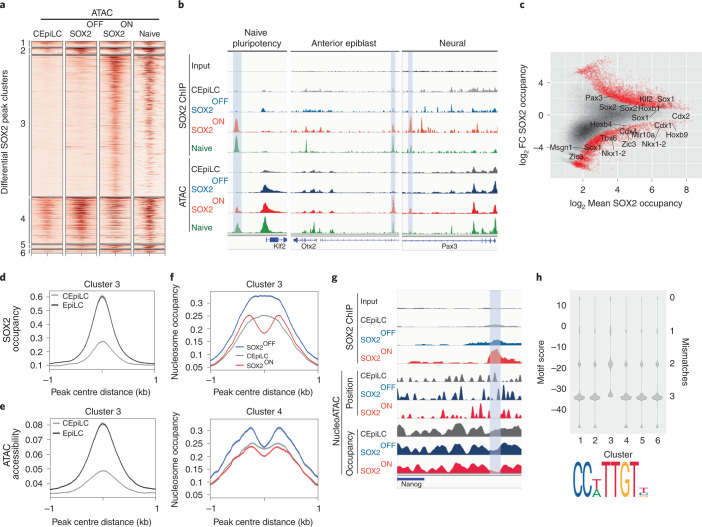
Fig. 5. SOX2 promotes chromatin accessibility at high-affinity sites.
a, Cluster 3 CREs classified in Fig. 3e exhibit ATAC–seq accessibility specifically in high-SOX2-expressing cells. Naive ATAC–seq data re-analysed from ref. 63. b, Correlated SOX2 occupancy and ATAC–seq accessibility at representative cluster 3 CREs (shaded blue) associated with genes expressed in high-SOX2-expressing cell types. c, SOX2 occupancy in CEpiLCs is higher at peaks associated with trunk patterning and mesoderm genes and lower at peaks associated with pluripotency and neural genes than in EpiLCs. Differentially occupied peaks (red) are statistically different between conditions (FDR <0.05). FDR was determined by DESeq2 padj metric from n = 3 biological replicates. d,e, Cluster 3 peaks exhibit greater average SOX2 occupancy (d) and ATAC–seq accessibility (e) in EpiLCs than in CEpiLCs (re-analysed from ref. 35). f, Average nucleosome occupancy at cluster 3 CREs is reduced at SOX2 peak centres in SOX2-ON, but relatively unchanged between cell types at cluster 4 peaks. g, Negative-correlation between SOX2 occupancy, nucleosome position and nucleosome occupancy at a representative CRE (shaded blue). h, SOX2 binding motifs at cluster 3 peaks most closely resemble the consensus sequence shown, as determined by FIMO calculated motif score (Methods). Source numerical data are available in source data.