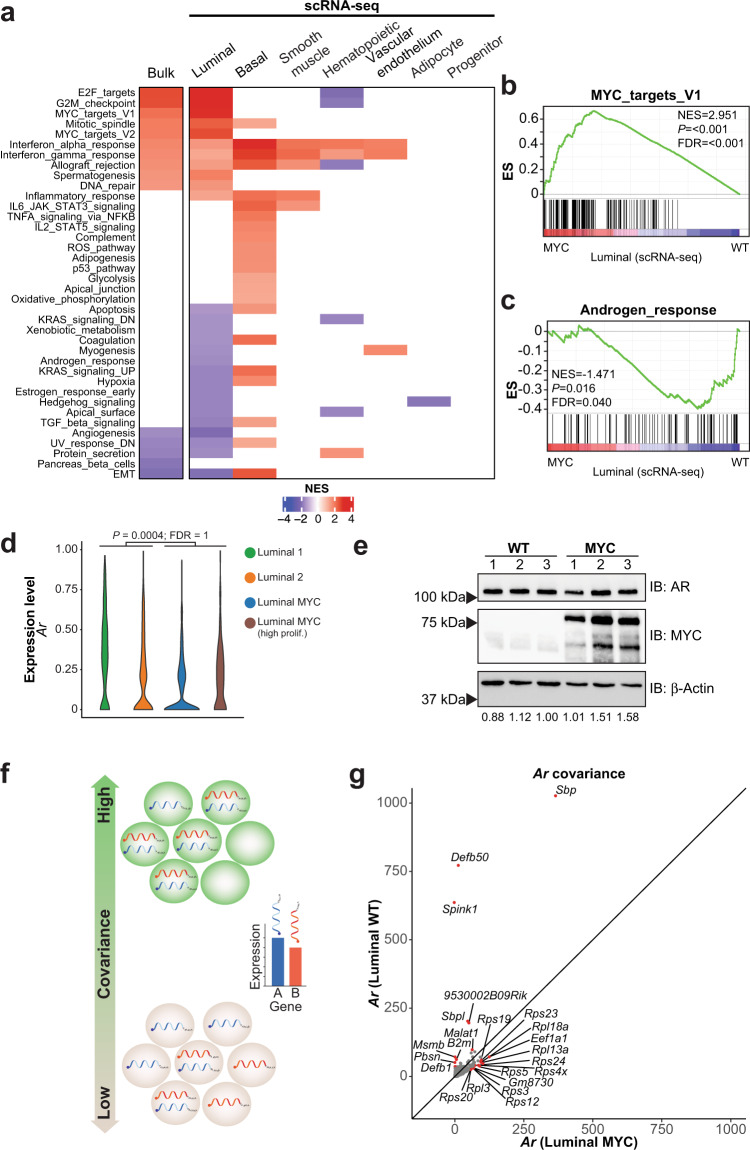
Fig. 3. MYC-driven luminal cells transformation dampens the AR transcriptional program.
a Gene Set Enrichment Analysis (GSEA, Hallmark, P < 0.05 and FDR < 0.1) revealed that the bulk RNA-seq transcriptional program associated with MYC overexpression is mostly driven by the luminal subset (VP; matched bulk and single-cell RNA-seq; n = 1 per genotype; Source data are provided as a Source Data file). b, c MYC overexpression is associated with an enriched MYC transcriptional program (b P < 0.001 and FDR < 0.001) and a depleted AR response (c P < 0.016 and FDR < 0.040) in the luminal subset (GSEA; VP; n = 1 per genotype). d, e MYC overexpression does not alter AR transcript expression in the luminal compartment (d VP; n = 1 per genotype; edgeR: two-sided quasi-likelihood F-test) and protein levels in the VP (e VP; n = 3 per genotype; numbers at the bottom represent AR levels relative to β-Actin; Source data are provided as a Source Data file). f Schematic representation of covariance analysis to determine co-expression (i.e. positive covariance) or mutually exclusive expression (i.e. negative covariance) between two genes at a single cell level. g Covariance analysis in the luminal subset reveals a shift from canonical AR target genes in the transcripts co-expressed with Ar upon MYC overexpression (VP; n = 1 per genotype). NES: normalized enrichment score; ES: enrichment score.