Fig. 6: Genetic contributions to centromere equalization in early embryogenesis.
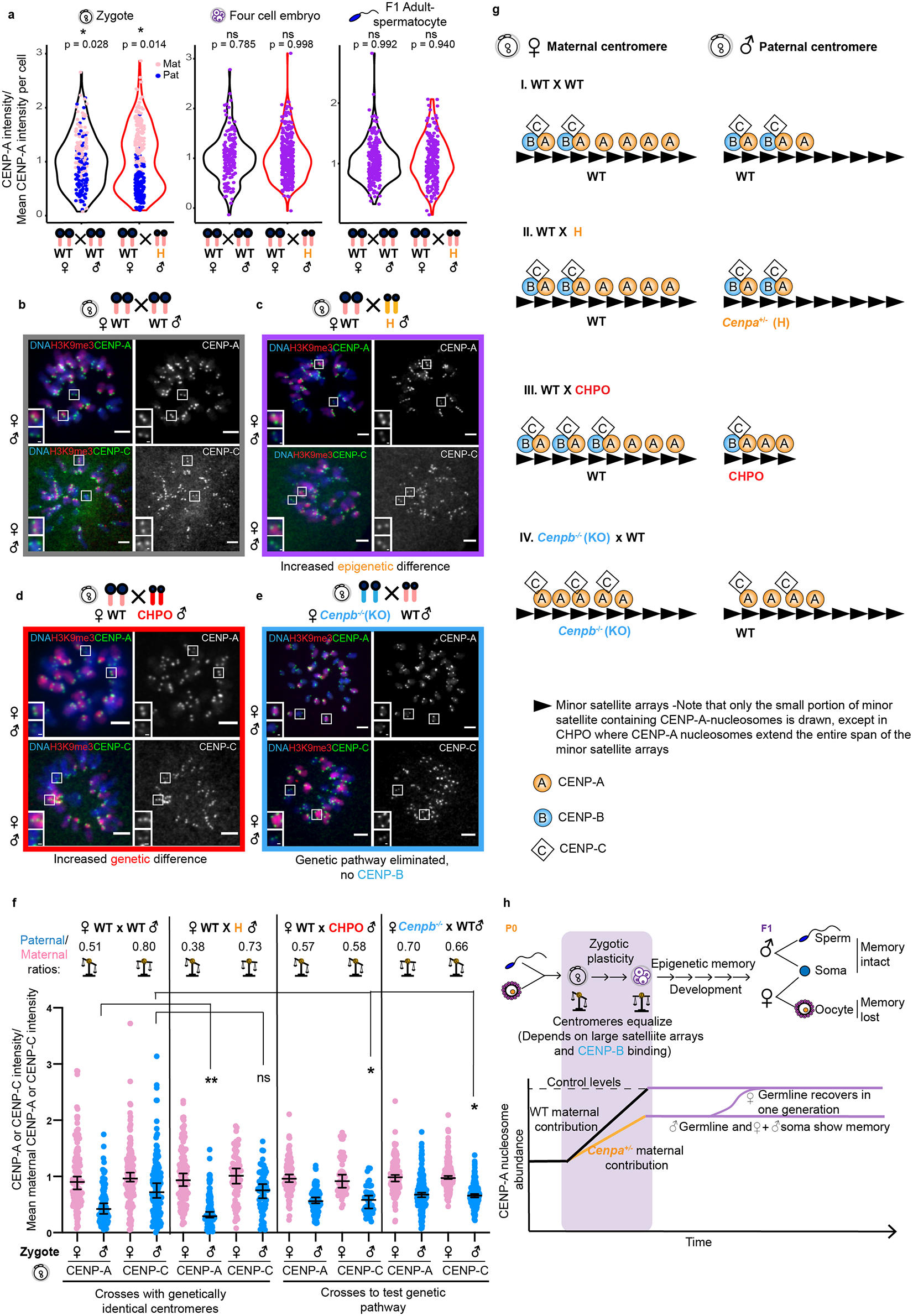
a, Combined violin and dot plots for zygotes, four cell embryos and adult spermatocytes showing the distributions of CENP-A intensities. Data for zygotes and spermatocytes are replotted from Fig. 3c and 1c, respectively. N = 192, 271, 164, 322, 240, 214 centromeres (left to right). Dot plots are colored for zygotes, where parental origin can be determined. *=‘modetest’52 for unimodality (two tailed). b-c, Images of CENP-A or CENP-C staining in zygotes with either moderate (WT × WT) or enhanced (WT♀ × H♂) epigenetic differences between maternal and paternal centromeres, distinguished by H3K9me3. Each pair of CENP-A or CENP-C foci represents sister centromeres in mitosis. d-e, Images of CENP-A or CENP-C staining in zygotes from the indicated crosses manipulating the genetic pathway. Scale bars: 5 μm (main panel), 1μm (inset). f, Quantifications of maternal (pink) and paternal (blue) CENP-A and CENP-C intensities in zygotes from the designated crosses, with average paternal/maternal CENP-A or CENP-C ratios above; N = 132, 90, 162, 157, 123, 112, 76, 66, 120, 116, 54, 45, 164, 172, 218, 217 centromeres (left to right). **/* P<0.0001/P< 0.05, Mann-Whitney U test (two tailed). Error bars: median ± 95% CI. g, Model for epigenetic and genetic contributions to CENP-C binding via CENP-A and CENP-B in zygotes. I. WT × WT cross: maternal centromeres have more CENP-A nucleosomes than paternal centromeres (Fig. 3c), but CENP-B is equally distributed between genetically identical maternal and paternal centromeres. We propose that CENP-B does not occupy all CENP-B boxes, and that CENP-C is limited relative to CENP-A and preferentially binds to CENP-A nucleosomes that are associated with CENP-B, thereby equalizing maternal and paternal centromeres. Note that only the small portion of minor satellite containing CENP-A-nucleosomes is drawn. II. WT × H cross: CENP-A nucleosomes are reduced on the paternal chromatin but still enough to recruit CENP-B/C. CENP-A asymmetry increases, but CENP-C remains symmetric. III. WT × CHPO cross: paternal CHPO centromeres have fewer minor satellite repeats and fewer CENP-B boxes. Most CENP-B therefore associates with maternal centromeres, providing more binding sites for CENP-C and increasing its asymmetry. IV. Cenpb−/− × WT: CENP-C binds to any available CENP-A nucleosomes, leading to CENP-C asymmetry matching CENP-A asymmetry. h, Summary of changes in centromeric chromatin at weakened paternal centromeres with WT zygotic genotype and either WT or reduced maternal contribution. See discussion. Note that weakened maternal centromeres would presumably lead to similar outcomes but are difficult to experimentally manipulate without also reducing the maternal contribution. Source numerical data are available in source data.
