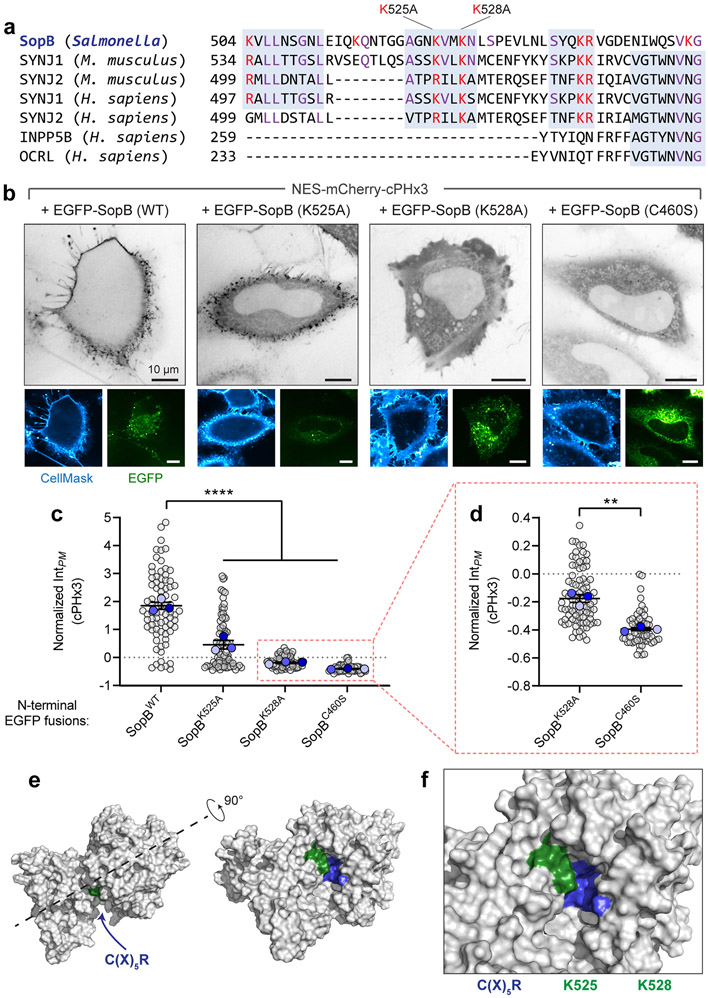
Extended Data Fig. 5. Lysine residues within the carboxyl-terminus of SopB support optimal phosphotransferase activity.
(a) Alignment of SopB amino acids 504-554 with other PtdIns(4,5)P2 and PtdIns(3,4,5)P3, 5-phosphatases. Basic residues in SopB are highlighted red along with conserved basic residues in the other phosphatases; additional conserved residues are highlighted purple, and regions of highest similarity are boxed (grey). Notably, residues that share sequence similarity with Mus musculus Synaptojanin-1 (SYNJ1) 534-584 fall outside of the SYNJ1 5-phosphatase domain and have not been implicated in its catalytic activity. Local homology between SopB and INPP5B or OCRL failed to be identified by NCBI BLAST.
(b) Mutation of lysines 525 and 528 blunt PtdIns(3,4)P2 generation by SopB. Wild-type, K525A, K528A, and C460S SopB amino-terminally tagged with EGFP (bottom panels) were transiently expressed with cPHx3 (inverted grey, main panels). The PM was stained with CellMask before imaging live. Representative confocal sections are presented for each. Note that expression of the K528A mutant led to notable perturbation of cellular morphology including rounding, cortical membrane ‘crumpling’, and blebbing.
(c) Normalized intensity of PM cPHx3 from (b) was quantified across n=3 independent experiments (WT, 78 cells; K525A, 81 cells; K528A, 86 cells; C460S, 57 cells). Data are trial means ± SEM (foreground) overlaid on cell measurements (gray, background). ****P < 0.0001.
(d) cPHx3 intensities from K528A- and C460S-SopB-transfected cells (red box, panel c) plotted on an expanded axis. Subtle PM-enrichment of cPHx3 was evident in K528A-transfected cells relative to the C460S-transfected mutant. Data are trial means ± SEM (foreground) overlaid on cell measurements (gray, background). **P = 0.0017.
(e,f) Lysine 525 and 528 of SopB are structurally predicted to neighbour the C(X)5R motif. The primary sequence of SopB (residues 1-561) was analyzed by RoseTTAFold and the resulting highest ranked model is presented in the surface filling view (grey). The location of lysine 525 and 528 (green) and the C(X)5R motif of SopB (blue) are annotated. Source numerical data are available in source data.