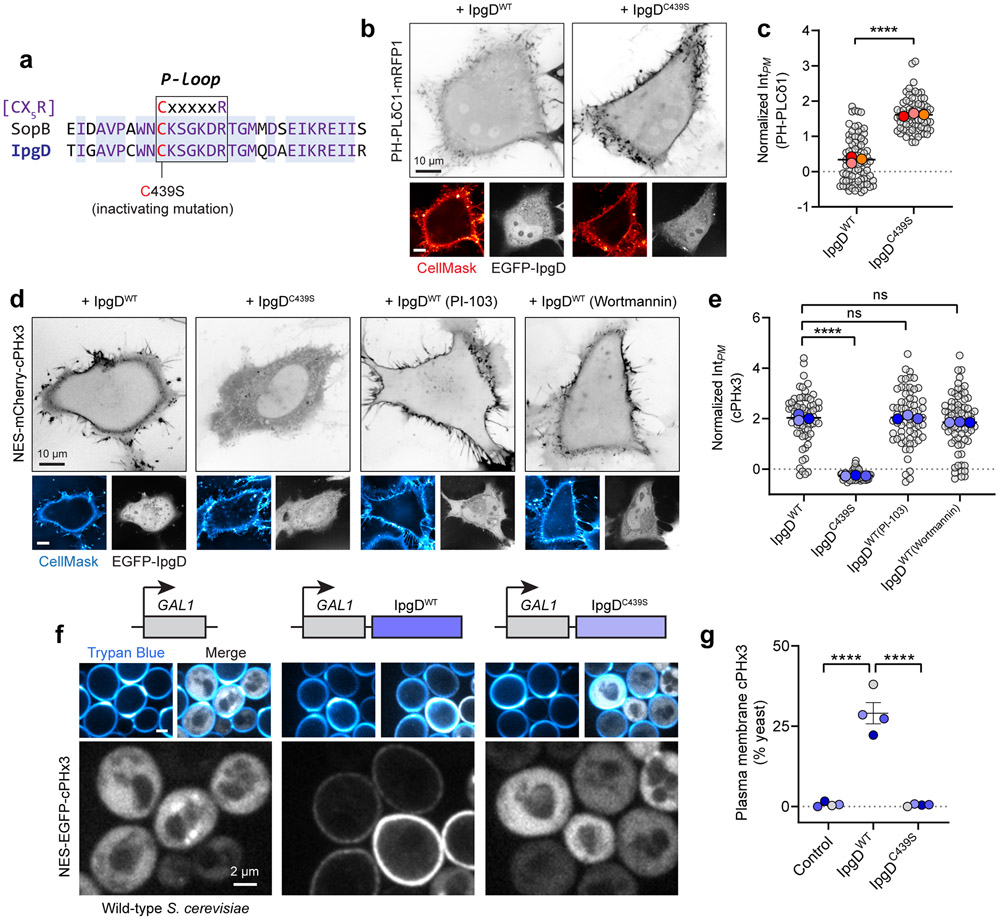
Extended Data Fig. 6. Generation of PtdIns(3,4)P2 by the Shigella flexneri effector IpgD does not require class I or class II PI3Ks.
(a) Sequence alignment of the phosphate-binding (P-loop) sequence from SopB (Salmonella enterica) and IpgD (Shigella flexneri). Residue 439 of IpgD encodes the cysteine of the C(X)5R motif.
(b) IpgD reduces PM PtdIns(4,5)P2 levels in a C(X)5R-dependent manner. Heterologous expression of EGFP-IpgD (WT or C439S) and PH-PLCδ1 (inverted gray, main panels). The PM was stained with CellMask prior to imaging live.
(c) Quantification of PH-PLCδ1 intensity in the PM from (b) across n=3 independent experiments quantifying 82 cells (IpgDWT) and 74 cells (IpgDC439S. Data are trial means ± SEM (foreground) overlaid on cell measurements (gray, background). ****P < 0.0001.
(d) Live cell imaging of heterologously-expressed EGFP-IpgD (WT or C439S) and NES-mCherry-cPHx3 following PM staining with CellMask. Cells were treated with the indicated inhibitors (PI-103, 500 nM; wortmannin, 100 nM) for 20 min prior to and throughout imaging.
(e) PM cPHx3 intensity was quantified from (d) across 3 independent trials (IpgDWT, 70 cells; IpgDC439S, 62 cells; IpgDWT(PI-103), 68 cells; IpgDWT(Wortmannin), 76 cells). Data are trial means ± SEM (foreground) overlaid on cell measurements (gray, background). ****P < 0.0001.
(f) PM PtdIns(3,4)P2 synthesis in S. cerevisiae. Galactose-inducible empty vector (control), IpgDWT, or IpgDC439S were induced for 2 hours in yeast that expressed cPHx3. Trypan Blue staining demarcates the cell wall and non-viable yeast.
(g) Yeast from (f) were scored for plasmalemmal cPHx3 localization across n=4 independent experiments analyzing the following number of yeast: control, 881 cells; IpgDWT, 1165 cells; IpgDC439S, 938 cells. Data are mean ± SEM. ****P < 0.0001. Source numerical data are available in source data.