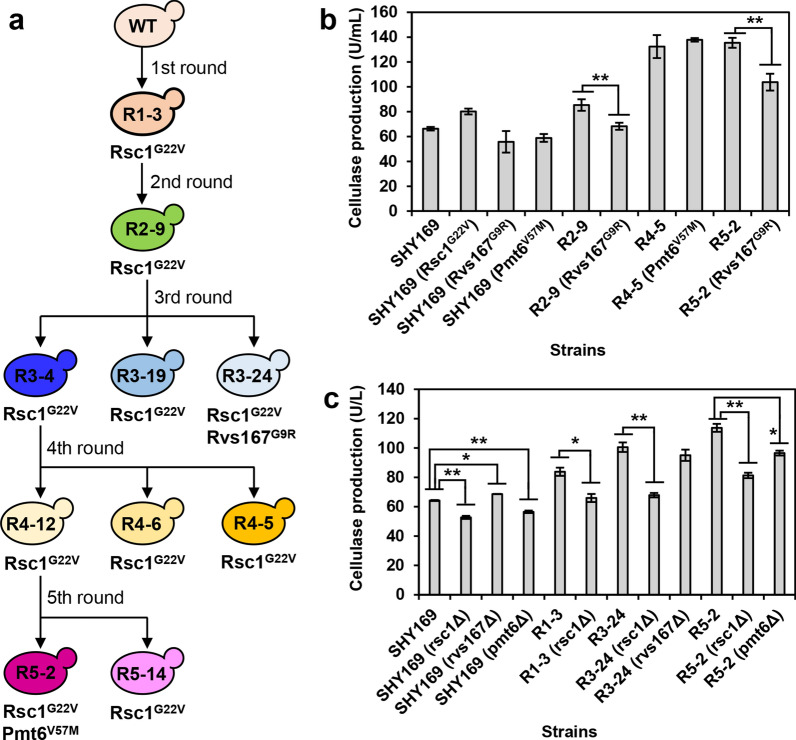
Fig. 7.
Identification of genomic variants using genome resequencing and genetic validation methods. a Protein point mutations uncovered by genome resequencing to be accumulated during the iterative ARTP mutagenesis and microfluidic screening. b Cellulase production in wild-type and reconstructed point mutation strains. c Cellulase production in wild-type and key gene deletion strains. Data represent the mean and standard error of duplicate cultures for each strain. Statistical analysis for each group of strains including the starting strain (SHY169, R1–3, R3–24, or R5–2) and its derived deletion strains was performed using one-way ANOVA followed by Tukey’s multiple-comparison posttest (**P < 0.01, *P < 0.05)