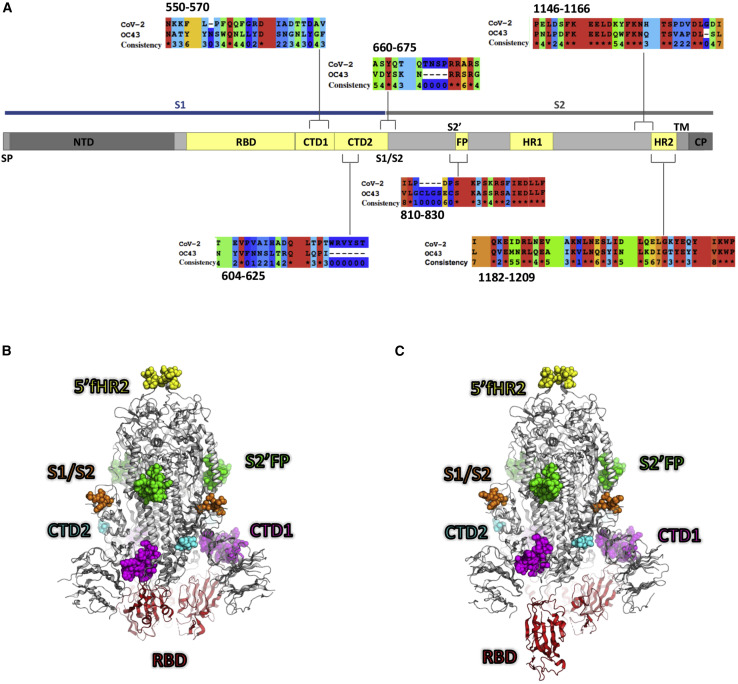
Figure 4.
Localization of SARS-CoV-2 spike immunodominant regions
(A) Diagram depicts SARS-CoV-2 spike protein subdomains, which include the N-terminal domain (NTD), the receptor-binding domain (RBD), S1-C terminus domains 1 and 2 (CTD1 and -2), the furin cleavage site (S1/S2); the S2′ cleavage site and fusion protein domain (S2′FP), the heptad repeat domains 1 and 2 (HR1 and -2), the transmembrane domain (TM), and the cytosolic domain (CP). Immunodominant regions with either high or low sequence identity with β-coronavirus OC43 are shown; asterisk indicates 100% sequence identity (red) and a consistency score of zero (blue) indicates no conservation of amino acid characteristics. (B and C) The homotrimeric SARS-CoV-2 spike protein is shown in the (B) closed and (C) open state (PDB: 6VXX and 6VYB, respectively). In each protomer of the spike, the protein mainchain is illustrated in white, except for the RBD, which is red. The atoms in the 6 peptide epitopes that were tested are shown as space-filling models, colored according to peptide number. There are regions of missing density in the models, presumably due to conformational flexibility, and these regions are omitted here; CTD1 (magenta) and S2′FP (green) are fully resolved; CTD2 (cyan), S1/S2 (orange), and 5′fHR2 (yellow) are partially resolved; and HR2 is completely absent in the structure.