Figure 4.
NLRP1 engages a caspase-3/Gasdermin E-dependent pyroptosis pathway upon SARS-CoV-2 infection
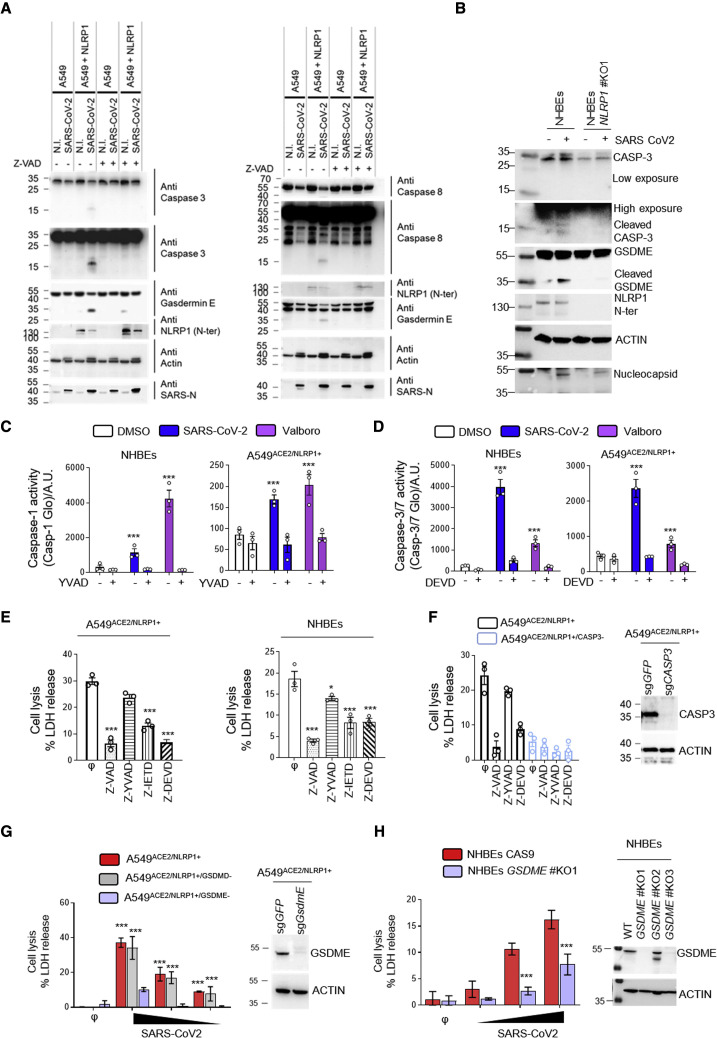
(A) Western blot examination of Gasdermin E, caspases-3, and caspases-8 processing in A549NLRP1+ and A549NLRP1− cells after 24 h of infection with SARS-CoV-2 (MOI 0.05) in the presence or absence of the pan-caspase inhibitor Z-VAD (25 μM). Immunoblots were performed against full-length and processed forms of Gasdermin E (p55 and p30), caspase-8 (p54 and p15), caspase-3 (p35 and p17/19), SARS-CoV-2 nucleocapsid (p40), NLRP1 N-terminal (p130/110), and ACTIN (p40).
(B) Western blot examination of Gasdermin E and caspases-3 processing in NHBEWT and NHBENLRP1−/− cells after 36 h of infection with SARS-CoV-2 (MOI 1). Immunoblots were performed against full-length and processed forms of Gasdermin E (p55 and p30), caspase-3 (p35 and p17/19), SARS-CoV-2 nucleocapsid (p40), NLRP1 N-terminal (p130/110), and ACTIN (p40).
(C and D) Measure of caspase-1 (C) and caspase-3/-7 (D) activities in SARS-CoV-2-infected (MOI 0.5) NHBEWT or A549NLRP1+ cells for 36 h in the presence or absence of inhibitors of caspase-1 (Z-YVAD, 40 μM) or caspase-3/-7 (Z-DEVD, 30μM). Val-boro (5 μM) was used a positive control of NLRP1-driven caspase activity for 10 H.
(E) Measure of cell lysis (LDH release) in A549NLRP1+ or NHBE-infected cells with SARS-CoV-2 (MOI 0.05 and 1, respectively) for 24 h in the presence/absence of the pan-caspase inhibitor Z-VAD (25 μM), the caspase-1 inhibitor Z-YVAD (40 μM), the caspase-8 inhibitor Z-IETD (40 μM), or the caspase-3 inhibitor Z-DEVD (30 μM).
(F) Western blot characterization of genetic invalidation of CASP3 in A549 population cells using CRISPR-Cas9 approaches and measure of cell lysis (LDH release) in A549NLRP1+ or A549NLRP1+/CASP3−-infected cells with SARS-CoV-2 (MOI 0.05) for 24 h in the presence/absence of the pan-caspase inhibitor Z-VAD (25 μM), the caspase-1 inhibitor Z-YVAD (40 μM), or the caspase-3 inhibitor Z-DEVD (30 μM). Efficiency of genetic invalidation by single-guide RNAs (sgRNAs) targeting GFP or caspase-3 was evaluated at the whole cell population.
(G) Western blot characterization of genetic invalidation of GSDME in A549 population cells using CRISPR-Cas9 approaches and measure of cell lysis (LDH release) in A549NLRP1+, A549NLRP1+/GSDMD−, or A549NLRP1+/GSDME−-infected cells with SARS-CoV-2 (MOIs 0.001, 0.01, and 0.1) for 24 h. Efficiency of genetic invalidation by single-guide RNAs (sgRNAs) targeting GFP or GSDME was evaluated at the whole cell population.
(H) Western blot characterization of genetic invalidation of GSDME in NHBE population cells using CRISPR-Cas9 approaches and measure of cell lysis (LDH release) in NHBEWT or NHBEGSDME−/−-infected cells with SARS-CoV-2 (MOIs 0.1, 0.5, and 1) for 36 h. Efficiency of genetic invalidation by single-guide RNAs (sgRNAs) targeting GFP or GSDME was evaluated at the whole cell population.
Data information: western blot (A, B, and E–G) images are from one experiment performed 3 times. Graphs (C–G) show data presented as means ± SEM from n = 3 independent pooled experiments; ∗∗∗p ≤ 0.001 for the indicated comparisons with t test.