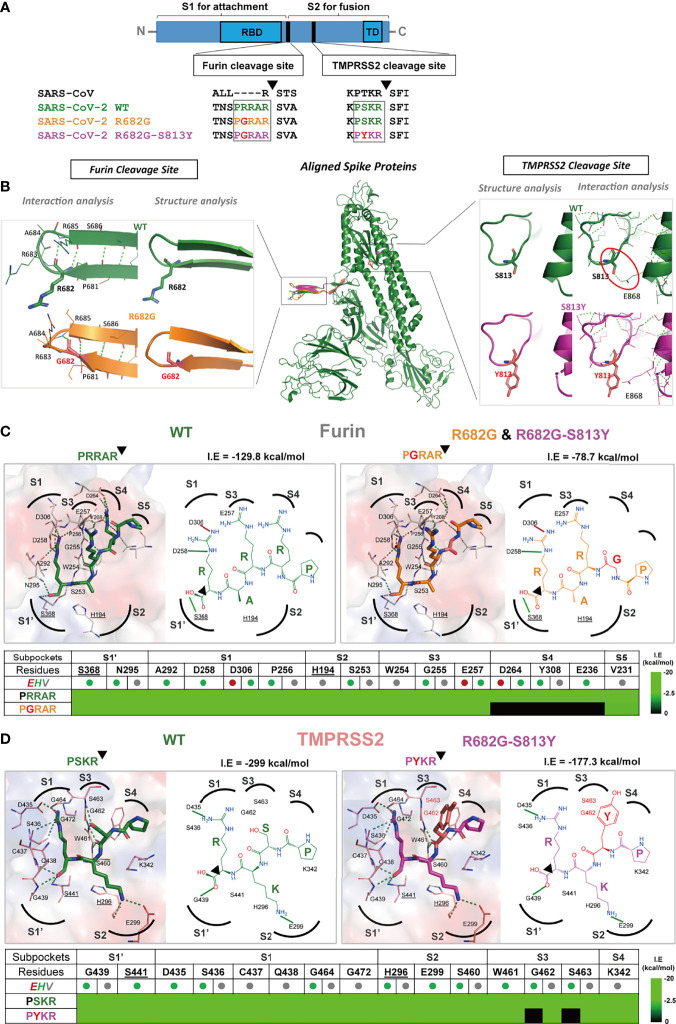
Figure 5.
The R682G and S813Y mutations in Spike protein lead to altered enzyme binding and proteolysis. (A) The Furin and TMPRSS2 cleavage motifs are shown in the spike protein sequences of SARS-CoV, SARS-CoV-2 WT, and mutants. Furin cleavage occurs between the S1 and S2 subunits, while the TMPRSS2 cleavage occurs within the S2 subunit. (B) The S protein structure and interactions of the WT (wild-type; green), R682G (orange), and R682G-S813Y (magenta) mutants were modeled using the Phyre2 web portal. The full spike proteins are aligned, and the specific mutation sites are shown in insights of the Furin and TMPRSS2 cleavage sites. (C, D) Molecular docking of spike protein peptide substrates (of WT and mutants) with Furin and TMPRSS2 using iGEMDOCK. The protease active site (surface) and the binding poses (2D diagrams) of the WT (green stick) and mutants (mutated residue in red sticks) were shown. The active site subpockets (dotted curves) with residues (catalytic residues labels underlined) were also displayed. The interactions (solid black lines, E-dark red, H-green, V-grey) and total interaction energies (I.E in kcal/mol) were examined. The interaction table details the substrate-subpocket residue interactions.