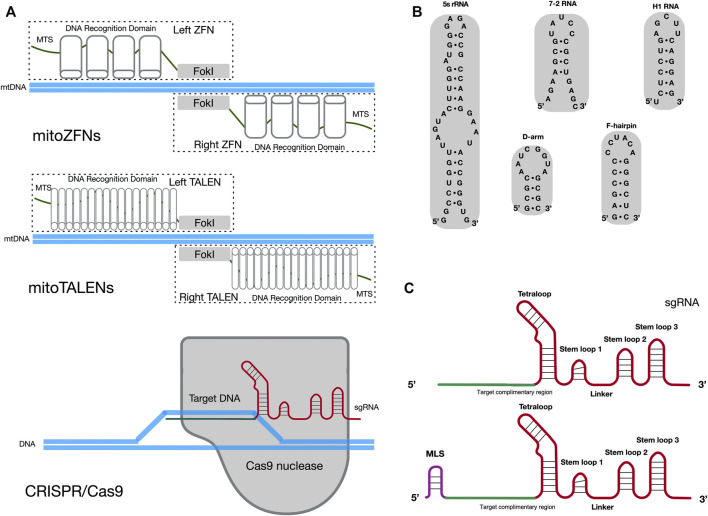
FIGURE 1.
Potential methods for sgRNA importation into mitochondria. (A) Diagram illustrating the cleavage mechanism of mitoZFNs, mitoTALENs and the CRISPR/Cas9 system. MTS, mitochondrial targeting sequence; FokI, non-specific FokI nuclease; sgRNA, single guide RNA. (B) Stem-loop motifs identified in human 5s rRNA, 7-2 RNA, H1 RNA,D-arm and F-hairpin motifs identified in yeast tRNALys (CUU). (C) The secondary structures of the original sgRNA and sgRNA with MLS. The original sgRNA is comprised of three stem loops and one tetraloop. When compared with the original sgRNA, the potential mitochondria-targeted sgRNA may be have an additional MLS at the 5′-end. The target complimentary region, which the RNA sequence for recognized target DNA, is shown in green. The backbone of the sgRNA is shown in red. MLS is the mitochondrial localization signal and is shown in purple.