Figure 1.
YTHDF2 is upregulated upon B cell activation prior to GC seeding
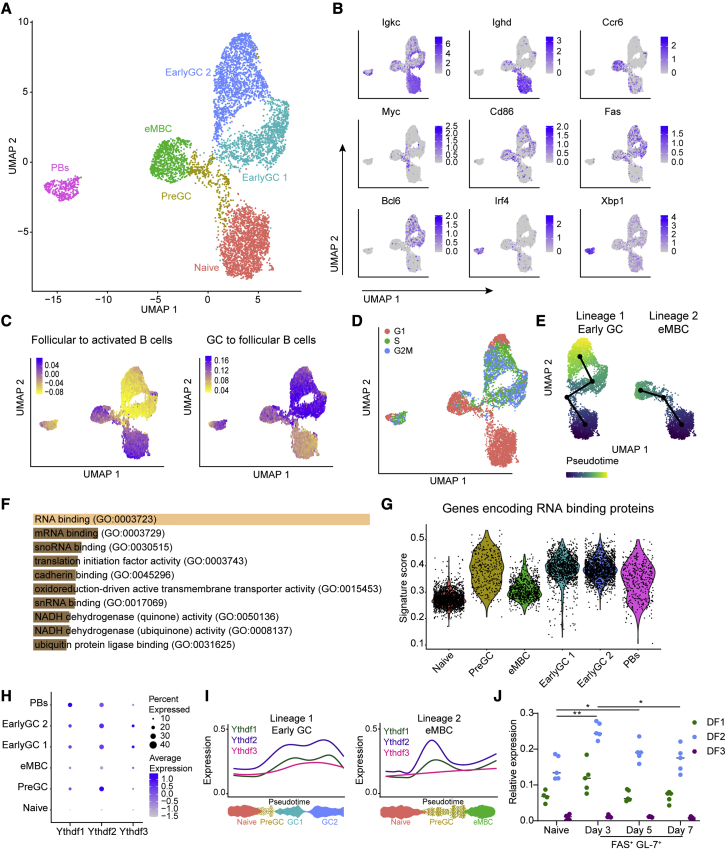
(A) Single-cell RNA-seq of transferred B1-8hi B cells pooled from three mice 5 days after immunization. UMAP projections of scRNA-seq profiles of 6,263 cells. Clusters in the UMAP plots are color coded according to different cell populations.
(B) Two-dimensional embedding as in (A) colored to represent the weight of genes associated with distinct cell clusters.
(C) UMAP projections of signature scores. Each signature consists of genes that are upregulated (fold change > 2, adjusted p < 0.05) in the indicated cell populations. Gene Expression Omnibus accession numbers GEO: GSE174394 and GSE60927 were used.
(D) Cell-cycle phase assignment of single cells based on gene expression.
(E) Pseudotime analysis of B cell differentiation trajectories analyzed using Slingshot R package.
(F) Molecular function GO terms enriched in genes whose expression is significantly upregulated in pre-GC compared with the naive and eMBC cell clusters. Terms are ordered based on p value ranking.
(G) Violin plots of the distribution of RBP gene signature scores in different cell clusters.
(H) Dot plot representation of Ythdf1/2/3 expression arranged by average expression and percent expression per cluster.
(I) Expression of Ythdf1/2/3 along the B cell differentiation trajectories defined in (B), (C), and Figure S1D.
(J) qRT-PCR of Ythdf1/2/3 expression in sorted FAS+ GL-7+ B1-8hi transferred B1-8hi B cells 3–7 days after immunization. Two to three primer pairs per gene were used to measure Ythdf expression. Plot shows average expression relative to a reference gene, Ubc. Data were pooled from five mice from two independent experiments. Lines indicate average mRNA expression. Statistical significance was tested by two-way ANOVA followed by Tukey’s multiple comparisons test. ∗p < 0.05, ∗∗p < 0.01; ns, not significant.