Figure 5.
The plasmablast genetic program is repressed by YTHDF2
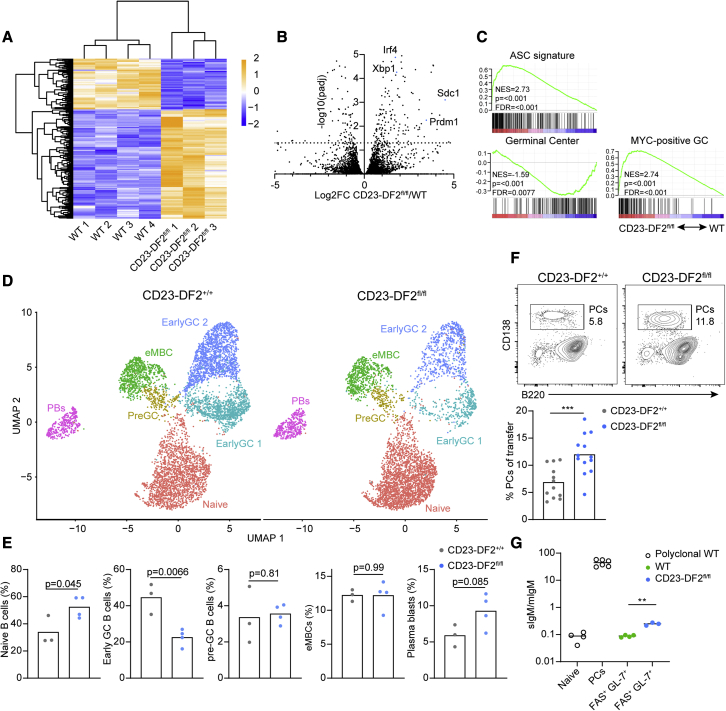
(A) Heatmap representation of clustering analysis of differentially expressed genes in transferred WT and CD23-DF2fl/fl FAS+ GL-7+ B1-8hi B cells sorted 5 days after immunization.
(B) Volcano plot of statistical significance against fold change, demonstrating the most significantly differentially expressed genes. PB-associated genes are marked in blue. Log2FC, Log2 fold change.
(C) GSEA of differentially expressed genes. Signatures were derived from public RNA-seq datasets (MYC-positive GC, GEO: GSE39443; GC, GEO: GSE110669; ASC, GEO: GSE60927).
(D) UMAP projections of scRNA-seq profiles of 6263 CD23-DF2+/+ and 4964 CD23-DF2fl/fl B1-8hi B cells 5 days after immunization. Data were pooled from 3–4 host mice per genotype. Clusters in the UMAP plots are color coded according to different cell populations. Annotation of cell populations was based on the same markers as in Figures 1B, 1C, and S1D.
(E) Quantification of scRNA-seq profiles per cluster based on Cell Hashing replicates.
(F) Analysis of PC formation potential of transferred B1-8hi B cells 5 days after NP-KLH immunization. Data were pooled from 12–14 mice from five independent experiments.
(G) qRT-PCR analysis of mIgM and sIgM mRNA transcripts derived from transferred WT and CD23-DF2fl/fl FAS+ GL-7+ B1-8hi B cells as in (A), as well as from polyclonal WT naive B cells and PCs. Data were pooled from a total of 3–6 mice. Lines indicate the mean.
Statistical significance was tested by two-tailed unpaired Student’s t test (E and F) or one-way ANOVA followed by Holm-Sidak multiple comparisons test (G). ∗∗∗p < 0.005.