Figure 4. Negative charges in the CHL are important for inhibiting stem RNA binding.
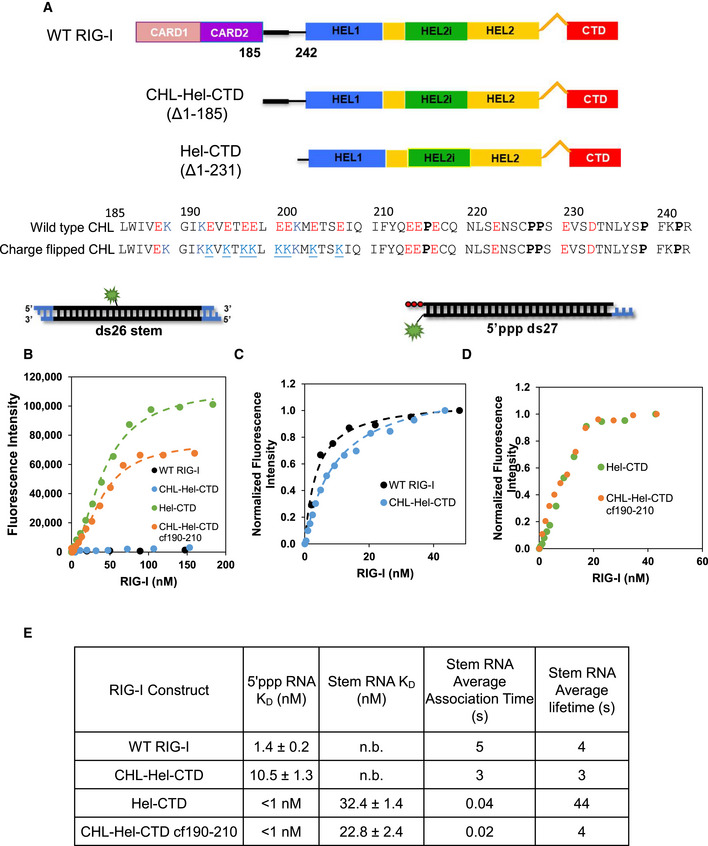
- Schematics of the tested RIG‐I constructs. The 190–210 CHL is highlighted by a thicker black bar. The negative amino acids are in red and positive ones in blue. The negatively charged amino acids in the 190–210 region were mutated to lysines to create the CHL‐Hel‐CTD cf190–210 construct. This change resulted in a pI change in the CHL from to 3.7 to 9.5.
- Fluorimetric titrations show ds26 stem RNA (Cy3 fluorophore at the 13th position) binding to the various RIG‐I constructs. The RNA binding data of Hel‐CTD and CHL‐Hel‐CTD cf190–210 were fit using a Hill equation (Hel‐CTD, n = 1.8 ± 0.2; CHL‐Hel‐CTD cf190–210 n = 1.9 ± 0.1). Because WT RIG‐I and CHL‐Hel‐CTD did not finish binding within 400 nM, they could not be normalized.
- Fluorimetric titrations show 5’ppp ds27 (DY547 at 3’ position distal to the 3nt DNA overhang) binding to WT RIG‐I and CHL‐Hel‐CTD. The fluorescence intensity changes were normalized to the highest fluorescence in each RNA. The data were fit using a hyperbola.
- Fluorimetric titrations show 5’ppp ds27 binding to Hel‐CTD and CHL‐Hel‐CTD cf190–210. Both proteins bind 5’ppp RNA stoichiometrically.
- Table summarizes the RNA KD values and shows the association and dissociation times of each RIG‐I construct. No binding or n. b. indicates that stem RNA binding was not observed within 400 nM of titrated RIG‐I. Each KD value was repeated twice (biological replicate, n = 2) The average stem RNA association and dissociation times of the various RIG‐I constructs from stopped‐flow experiments (Fig EV4 and Appendix Table S2).
