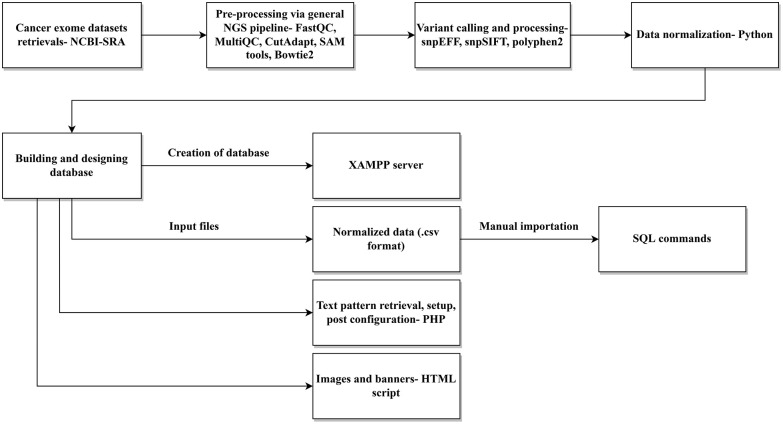
Figure 1.
Schematic representation of the MutaXome workflow. An initial experimental analysis of retrieved cancer exomes, including preprocessing, variant calling, and data normalization, revealed the different mutations in .csv format, which was used as the input for developing the database. MutaXome was created using the XAMPP server. The retrieval of text patterns, setup, and post configuration was carried out using PHP and the images and banners were added via HTML scripts. The final designed database can be installed and run as stated in the article.