Figure 1.
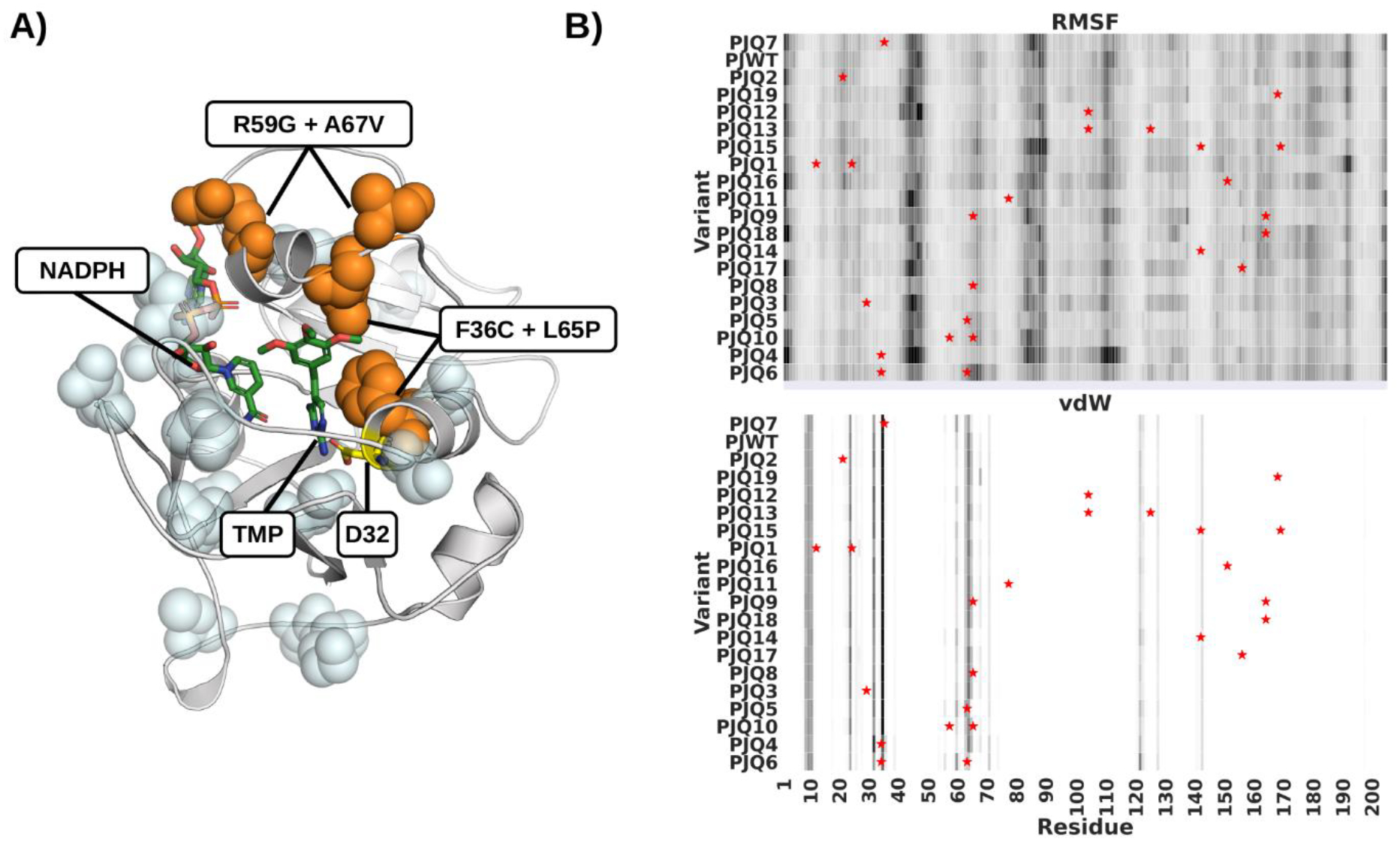
Mutant Variants of pjDHFR. A) Homology model of pjDHFR in complex with NADPH and TMP. Amino acid substitutions that lead to a minor (<10 fold) decrease in TMP binding are highlighted in light blue. Substitutions that lead to a >10 fold decrease in TMP binding are labeled and highlighted in orange. B) Heatmap of pjDHFR C-alpha root-mean-squared fluctuations and vdW interactions with TMP calculated from the MD simulations. Rows presents variants, columns residues, darker colors indicate higher fluctuations and stronger vdW interactions. Variants are sorted by TMP Ki in descending order. Sites of mutations are marked by stars.
