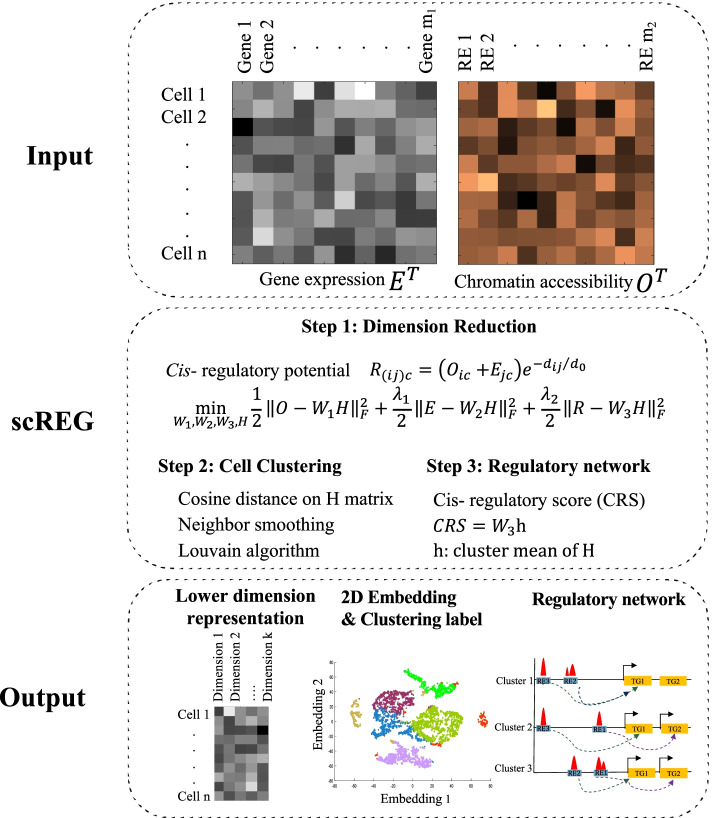
Fig. 1.
Schematic overview of the scREG. The scREG takes matrices of gene expression and chromatin accessibility (E and O respectively) measured on the same cells as inputs and process the multiome data by three steps: dimension reduction, cell clustering, and regulatory network inference. First, a cell level index cis- regulatory potential matrix R is defined, indicating the regulatory strength of a peak to a target gene. Rows of R matrix represent preselected peak-gene pairs and columns represent cells. Here, the index i represent the ith peak, j represents a gene, and c represents a cell. Then, three matrices (E, O, and R) are factorized into products of domain specific profiles (W1, W2, and W3 for E, O, and R respectively) and a common low dimension representation of the cells (the H matrix) by NMF-based optimization model. Based on the reduced dimension matrix, we do cell clustering by Louvain algorithm and visualize the cells into 2D space by Umap. For each peak-gene pair, a cis- regulatory score (CRS) was defined by average of the cis- regulatory potential over cells from the same cluster. Subpopulation specific cis- regulatory networks were identified based on the CRS scores.