Fig. 4. Context-dependent coding of subsequences in ALM.
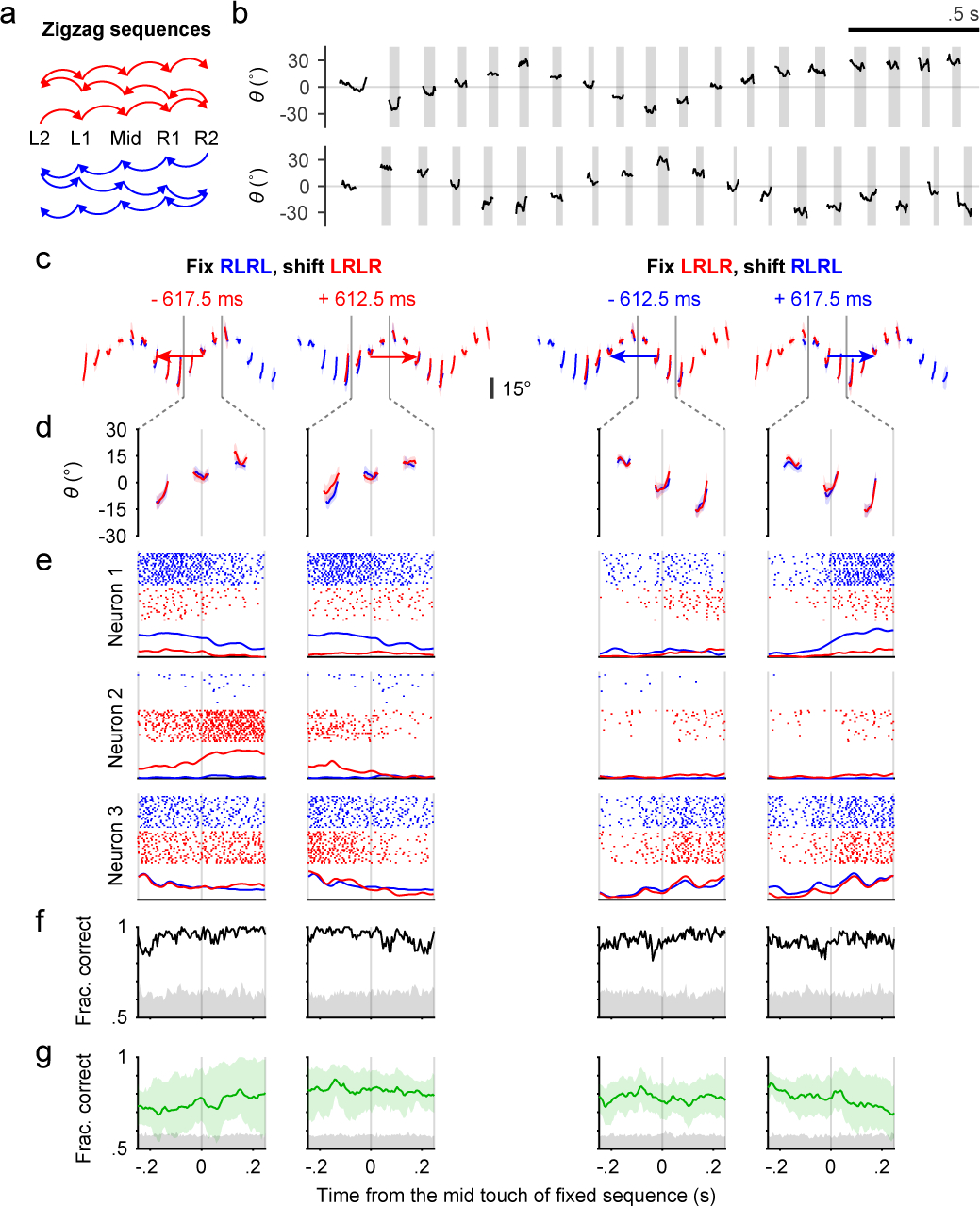
a, Transition diagrams depicting the two “zigzag” sequences, which contain symmetrical transitions.
b, Example trials showing patterns of tongue angle in the two “zigzag” sequences.
c, The four ways to shift and match subsequences. Colored traces show tongue angles from an example session (mean ± SD). Arrow colors indicate the sequence to be shifted. Arrow lengths and the number in milliseconds shows how much the chosen sequence must be shifted in order to match the other. Two gray vertical bars indicate the time window for analysis.
d, Zoomed-in plots of (c) showing the three licks in the middle of matched subsequences.
e, Example rasters and PETHs for three simultaneously recorded neurons. PETHs are normalized to each neuron’s maximum spike rate across the four shifts.
f, Classification accuracy (black trace) for sequence identity based on population activity for the session in (c-e). Chance accuracy (gray shading) was determined by randomly shuffling sequence labels.
g, Similar to (f) but showing mean ± 95% hierarchical bootstrap confidence interval across sessions (n = 6) and mice (n = 3).
