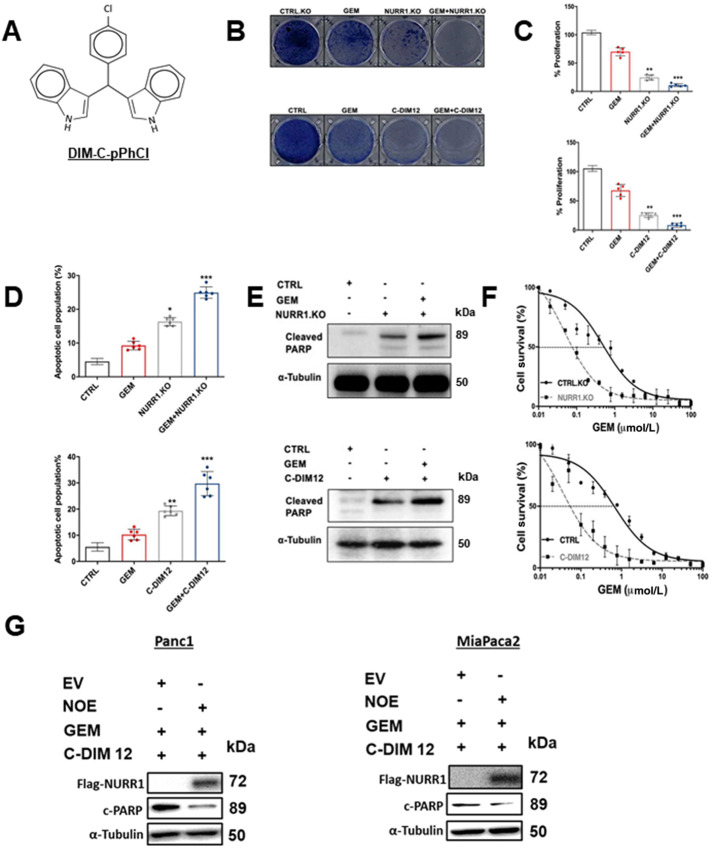
FIGURE 2.
NURR1 plays a cytoprotective role in pancreatic cancer. A, Chemical structure of DIM-C-pPhCl (C-DIM12). B, Images of crystal violet stained dishes that were treated with vehicle control, gemcitabine (GEM), C-DIM12, or NURR1 loss for 10 days after plating 1,000 cells per well in a 12-well plate (three independent experiments performed). C, Quantification of crystal violet staining is shown. Error bars indicate SEM of triplicate wells from a representative experiment (three independent experiments performed; **, P < 0.01; ***, P < 0.001). D, Apoptotic cell fraction was determined after treatment control (vehicle or CTRL.KO), gemcitabine, C-DIM12 or NURR1 loss, or a combination of both (gemcitabine + C-DIM12/ gemcitabine +NuRR1.KO for 72 hours. Apoptotic cell death was quantified by Annexin V/propidium iodide (PI) staining and flow cytometry and is shown as the percentage of cells that were PI positive. Each data point represents the mean ± SEM of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001). E, Immunoblot analysis of cleaved PARP in control, gemcitabine, C-DIM12, or NURR1 loss treated cells with α-tubulin as a loading control in MiaPaCa2. F, Drug sensitivity was measured by PicoGreen DNA quantitation, in MiaPaCa2 cells, under the indicated culture conditions, and with varying doses of gemcitabine. Each data point represents the mean of four independent measurements. G, Panc1 MiaPaca2 cells were treated as indicated in the presence or absence of NOE or expression of empty vector (EV) and whole-cell lysates were analyzed by Western blots as outlined in the Materials and Methods.