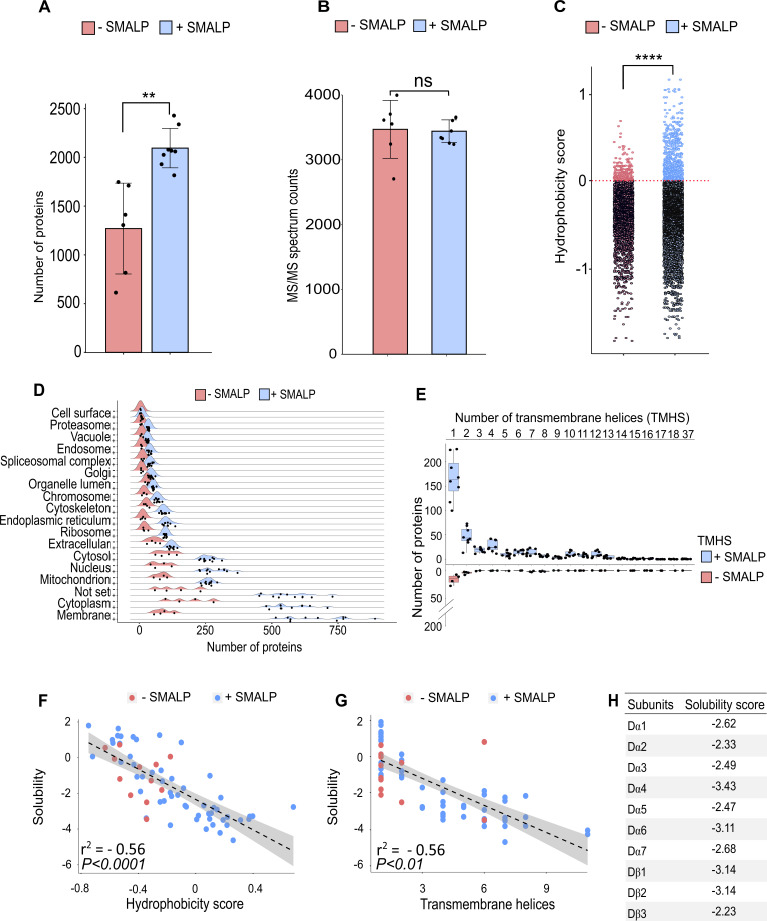
Figure 4. Identification of proteins enriched by SMALP extraction.
(A) Number of identified proteins in affinity pull-down samples solubilised with or without SMA, two-tailed t-test, **p<0.01, n=6 or 8 replicates per condition. (B) MS/MS spectrum counts from samples solubilised with or without SMA, ns = not significant after two-tailed t-test with n=6 or 8. (C) Calculated hydrophobicity score of amino acid residues found in protein sequences obtained with and without SMA solubilisation, ****p<0.0001, two-tailed t-test, n=3 per condition. (D) GO term (cellular compartment) enrichment of proteins identified with and without SMA solubilisation, n=4 or 11. (E) Predicted numbers of proteins containing transmembrane helices obtained with or without SMA solubilisation, n=4 or 8. (F, G) Analysis of solubility and hydrophobicity of receptors identified with and without SMA solubilisation (r2 = –0.56, p<0.0001, n=4) and of transmembrane receptor helices (r2=0.56, p < 0.01, n = 4). (H) Solubility score of individual nAChR subunits.