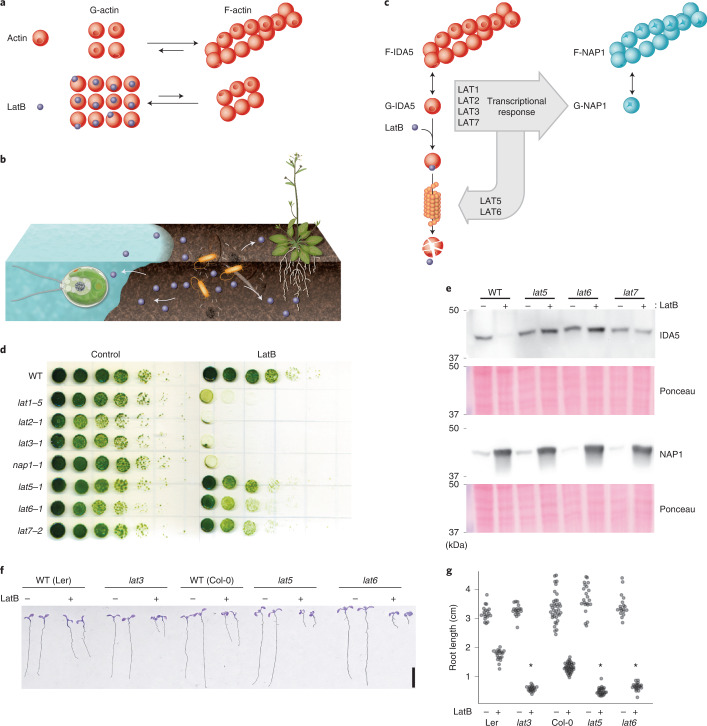
Fig. 5. The approach revealed new conserved components of a defense mechanism against cytoskeleton inhibitors.
a, LatB interferes with actin polymerization. b, Soil microorganisms deploy (arrows) actin inhibitors (blue circles) for a competitive advantage in their environment. c, Chlamydomonas responds to actin inhibition by degrading its conventional actin, IDA5, and upregulating an alternative actin, NAP1. d, Growth of new lat mutants identified in this study (lat5-1, lat6-1 and lat7-2) was compared to previously isolated lat1-5, lat2-1, lat3-1 and nap1-1 mutants66 in the absence (control) and presence (LatB) of 3 µM LatB. e, Immunoblot of conventional (IDA5) and alternative (NAP1) actins shows that lat5-1, lat6-1 and lat7-2 are deficient in actin degradation. Immunoblot representative of n = 3 independent experiments. f, The F-actin homeostasis pathway is conserved between green algae and plants. Mutants in Arabidopsis genes homologous to Chlamydomonas lat3, lat5 and lat6 are sensitive to LatB, as evidenced by decreased root length. g, Quantification of root length in Arabidopsis mutants. Asterisks mark significant changes relative to wild type under the same condition based on two-way analysis of variance. The exact value of P = 2.4 × 10–47 (Ler versus lat3), P = 1.4 × 10–62 (Col-0 versus lat5), P = 6.8 × 10–23 (Col-0 versus lat6). n = 26 roots examined over three independent experiments.