Fig. 2. Population GWAS estimate the association between raw genotypes G and phenotypes X.
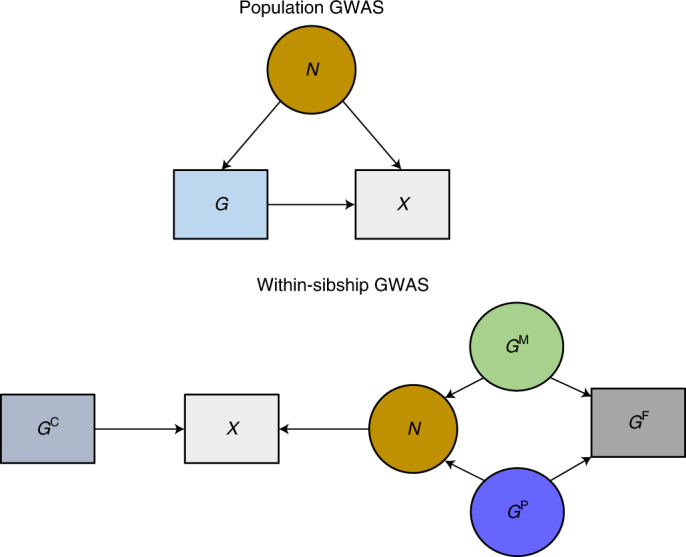
As outlined in Fig. 1, estimates from population GWAS may not fully control for demography (population stratification and assortative mating) and may also capture indirect genetic effects of relatives. For simplicity we use N to represent all sources of associations between G and X that do not relate to direct effects of G. Circles indicate unmeasured variables and squares indicate measured variables. If parental genotypes are known, G can be separated into nonrandom (determined by parental genotypes) and random (relating to segregation at meiosis) components. Within-sibship GWAS include the mean genotype across a sibship (GF) (a proxy for the mean of the paternal and maternal genotypes GP, M) as a covariate to capture associations between G and X relating to parents. The within-sibship estimate is defined as the effect of the random component: that is, the association between family-mean-centered genotype GC (that is, G − GF) and X. Demography and indirect genetic effects of parents (N) will be captured by GF. The association between GC and X will not be influenced by these sources of association but could be affected by indirect effects of the siblings themselves, which are not controlled for.
